| Posted: Feb 14, 2008 | |
Protein engineering - from the humble spider to the nanotechnology future of material design |
|
| (Nanowerk Spotlight) Much has been written about the fascinating properties of spider silk, a biopolymer that is stronger than steel and more elastic than rubber. The silken threads possess a unique combination of mechanical properties: strength (its tensile strength is about five times as strong a steel of the same density), extensibility (up to 30%) and toughness (its ability to absorb a large amount of energy without breaking). Of course this begs the obvious question: How is it possible that spider silk - produced by little creatures that evolved about 400 million years ago - can be as strong as steel - a modern alloy that plays a critical role in our infrastructure and which still attracts considerable R&D investments in its production technology? | |
| What is perplexing is that the atomic interactions (hydrogen- or H-bonds) in spider silk are actually 100 to 1,000 times weaker than those in steel, or than those in the super-fiber Kevlar, where covalent bonds are used. Hydrogen bonds are the basic chemical bonds that hold together proteins, similar to trusses and beams in buildings, and play a key role in controlling the behavior of these structures. In order to reach silk's mechanical properties, most synthetic materials must be much denser, thus much heavier and consume much more energy during their synthesis and transport. | |
| New analysis performed at MIT’s Laboratory for Atomistic and Molecular Mechanics shows that the intriguing strength of spider silk may be made possible by precisely controlling the number and the geometry of H-bonds at a characteristic length scale. The physical concept is that by making many small elements work together cooperatively, the weaknesses of the individual components can be overcome. All this must happen at the nanoscale in order to be effective. | |
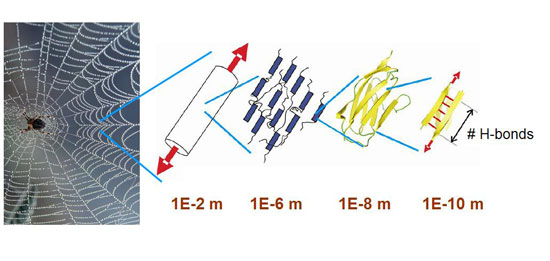 |
|
| The extraordinary mechanical properties of spider silk may be made possible through precisely controlled number and geometry of H-bonds at a characteristic length scale. (Image: Dr. Buehler, MIT) | |
| Despite scientists' significant advancements in their understanding of the nanomechanics of biological materials, several key fundamental questions remains unanswered: What is the strength limit of H-bond assemblies, and how is it possible that protein materials such as spider silk reach strengths that exceed those of steel, despite the weakness of H-bond interactions? The mechanical behavior of protein materials is exceedingly complex, partly due to the hierarchical architecture of these materials that extends from nano- to macroscale. Unraveling the mysteries of Nature's protein design blueprints will have a big impact on the future of nanotechnology applications in materials design and engineering. | |
| "Our recent findings explain how the intrinsic strength limitation of H-bonds in spider silk is overcome by the formation of a nanocomposite structure of H-bond clusters, thereby enabling the formation of larger and much stronger beta-sheet structures" Prof. Markus Buehler explains to Nanowerk. "We have discovered this by bringing together two independent concepts that were established earlier in the field of proteins (both are universal properties of proteins, that is, they do not depend specifically on which protein is considered, and both have been clearly confirmed in experiment): | |
| "1. Proteins are highly flexible structures that easily bend and turn, leading to a particular form of elasticity called “entropic elasticity”. Many experiments with AFM have shown that this 'entropic elasticity' is indeed the governing mode of deformation. | |
| "2. Proteins are bound together by the well-known H-bonds (one of the weakest chemical bonds known). It is established that these H-bonds contain very little energy. In fact, their energy is so small that they can easily break just by the random thermal vibrations at the molecular level if they don’t work cooperatively in assemblies." | |
| Earlier studies were unable to combine these two properties of protein structures in elucidating their strength. The 'entropic elasticity' alone, or the properties of H-bonds alone can not explain how spider silk can be as strong as steel. | |
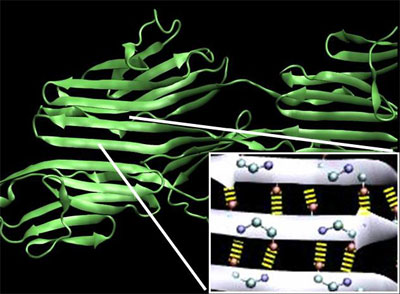 |
|
| Molecular structure and loading conditions used for molecular dynamics simulations. Beta-sandwich structures are a characteristic feature of mechanical proteins, which employ networks of parallel H-bonds. Interstrand H-bonds (thick yellow lines in insert) act as mechanical clamps that resist unfolding. (Image: Dr. Buehler, MIT) | |
| Buehler's group has achieved this link between the two by returning to a more fundamental level, using thermodynamics as the basic principle. | |
| "Interestingly, the same thermodynamical approach has been used more than 80 years ago during the development of the theories of fracture mechanics – at that time, applied to ships, and thus far used primarily at much larger scales" says Buehler. "In a different form but with essentially the same physics, it can also be applied to predict the fracture of proteins." | |
| Sinan Keten, an MIT graduate student, and Markus Buehler, the Esther and Harold E. Edgerton Professor at MIT's Department of Civil and Environmental Engineering, and his group focus on the systematic understanding of the macroscopic mechanical response of complex materials based on their fundamental, atomistic ultrastructure using large-scale, massively parallelized novel modeling techniques that provide an atom-by-atom view of the rupture events in protein materials. We already reported on some of his fascinating previous work: "Nanostructure will become design variable for materials engineers" and "Unlocking nature's protein material concepts enables tomorrow's nanotechnology materials and cures". | |
| By working out this new model, which has been reported in the February 15, 2008 online edition of Nano Letters ("Geometric Confinement Governs the Rupture Strength of H-bond Assemblies at a Critical Length Scale"), Buehler and Keten made a surprising discovery: H-bonds in proteins are special atomic bonds with fascinating properties. | |
| He explains that using only one or two H-bonds in building a protein provides no or very little mechanical resistance. Using three to four H-bonds, however, leads to a resistance that actually exceeds that of many metals. Using more than four bonds also leads to a much reduced resistance. This means that the strength is maximized at a characteristic number of H-bonds. | |
| "We have not only performed a theoretical and simulation analysis of this phenomenon" says Buehler. "We have also cross-checked our results against a variety of experimental observations from different sources and different materials. Among others, we utilized a large experimental database with thousands of protein structures (from the Protein Data Bank) and analyzed the prevalence of the characteristic 'size' of the protein, focused on beta-sheet rich protein structures (these are structures that are often found in structural protein materials). What we found was quite exciting: The above described strength resistance as a function of size correlates very closely with the biological prevalence." | |
| This suggests that a key evolutionary driving force for the structural features of these proteins may be related to strength: Protein structures at a characteristic dimension are most common in biology. Protein structures smaller or larger than this characteristic dimension are less common. | |
| Buehler explains how these findings are important for several reasons: | |
| 1) It may help us to understand what type of protein mutations lead to malfunctions of the protein, for instance in genetic diseases that disrupt the H-bonding structures or those that create overly strong structures. | |
| 2) Since H-bonds are an important structural 'glue' (in some sense, the cement of biology), and since mechanical forces play a critical role in many biological processes, the findings could help us to better understand how elementary biological processes work (e.g. mechanotransduction). | |
| 3) It may enable us to design new materials based on proteins. It is already possible to synthesize novel materials from scratch, using protein elements as building blocks. What is missing are engineering tools that direct this material design process. | |
| 4) The findings may be important to better understand the behavior of other protein materials, for instance those that define hair, protein nanotubes and amyloids, muscle proteins, or those that make up the cell’s cytoskeleton. | |
 By
Michael
Berger
– Michael is author of three books by the Royal Society of Chemistry:
Nano-Society: Pushing the Boundaries of Technology,
Nanotechnology: The Future is Tiny, and
Nanoengineering: The Skills and Tools Making Technology Invisible
Copyright ©
Nanowerk LLC
By
Michael
Berger
– Michael is author of three books by the Royal Society of Chemistry:
Nano-Society: Pushing the Boundaries of Technology,
Nanotechnology: The Future is Tiny, and
Nanoengineering: The Skills and Tools Making Technology Invisible
Copyright ©
Nanowerk LLC
|
Become a Spotlight guest author! Join our large and growing group of guest contributors. Have you just published a scientific paper or have other exciting developments to share with the nanotechnology community? Here is how to publish on nanowerk.com.
