| Posted: Oct 10, 2017 | |||||||
Hydrogen bond strength in DNA: where 3 could be less than 2 |
|||||||
| (Nanowerk Spotlight) Hydrogen bond base pairing forces are essential for the mechanisms associated with DNA stability. Despite attracting great research attention, this fundamental interaction has eluded a precise physical description so far since its electrical origin has not been quantified yet. | |||||||
| The general understanding of hydrogen bonds even today is limited to general structural and thermodynamic parameters (read more: "Defining the hydrogen bond: An account"). | |||||||
| Researchers now have proposed characterization by means of electrical forces, providing a framework for universal characterization of hydrogen bonds. In this way, they provide technical arguments to support that hydrogen bonds are well distinguishable and their role in biological events require a proper specific intrabond description. | |||||||
| Published in PLOS One ("Unveiled electric profiles within hydrogen bonds suggest DNA base pairs with similar bond strengths"). | |||||||
| This is the first report on the electrical characterization of DNA with intrabond resolution. | |||||||
| Specifically, it quantifies electrical forces due to a single hydrogen bond in DNA and provides proof of a non-direct relationship between the number of hydrogen bonds in DNA pairs and the relative strengths of such pairs. | |||||||
| Such understanding of the relative strengths of the forces involved in the specific bonding of DNA as well as its electrical origin provides physical foundations to control mechanisms associated with DNA stability. It could help to develop new methods for the understanding, characterization; and control of relevant events originated at intrabond scales e.g. controlled DNA repair and damage; controlled modification of the expression of the genome; and communications below the single bond limit. | |||||||
 |
|||||||
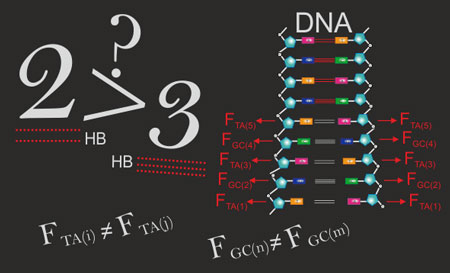
| A blackboard representation of the manuscript's key message: the quantification of the relative strengths between base pairs in DNA due to zipping hydrogen bonds might place on doubt such mechanisms regarding the interpretation of thermodynamic properties of DNA based on the assumption that A/T pairs are weaker than G/C pairs due to the sole difference in the number of hydrogen bonds, 2 and 3 respectively. (Image: Dr. Yamila García-Martínez) | |||||||
| Generally, being able to control DNA stability at the single bond level by means of electromagnetic interactions opens new avenues to induce modifications of the replication and transcription processes of DNA by means of noncontact methods. | |||||||
| "Our approach should permit a deeper knowledge of fundamental mechanisms occurring down to intrabond level, here with an initial focus on base-pair strengths," Yamila García-Martínez, senior author of the paper, tells Nanowerk. "It facilitates the reinterpretation of electrical features of hydrogen bonds in DNA and how it could lead to applying similar research and models to other life-essential molecules and events." | |||||||
| "Fundamentally, every chemical interaction relies mainly on the electrical forces among its constituent particles – electrons and nuclei," Yasser Bruno Ruiz-Blanco, the paper's first author, adds. "The electric field is a classic and easily tuneable property in the macro and micro world. Therefore, this work would contribute to pave the way to obtain the same capabilities of electric properties in the nano-world." | |||||||
| These results could contribute to understand previous controversial results regarding the role of hydrogen bonds in the electromechanical response of DNA. | |||||||
| "We issue an indicative warning in regards to the fact that 3 hydrogen bonds in the G/C base pair do not necessarily provide a stronger interaction than the 2 hydrogen bonds in A/T pairs as is being interpreted from some previous larger-scale experimental results," says García-Martínez. "In the case of DNA events, limitations as such related with the understanding of biological events attending to the sole number of hydrogen bonds should be place on doubt." | |||||||
| The table below include some of the published works that, according to the scientists, would be reviewed by results such as presented by them: | |||||||
|
|||||||
| Going forward, the researchers will study the effects of external electromagnetic fields on DNA at the level of single bond events. This will have not only an enormous interest in the medical field but also in nanotechnology where it would open the door to non-contact atomic manipulation of DNA – the analogue to the CRISPR gene editing method but using electromagnetic fields to drive changes in DNA. | |||||||
 By
Michael
Berger
– Michael is author of three books by the Royal Society of Chemistry:
Nano-Society: Pushing the Boundaries of Technology,
Nanotechnology: The Future is Tiny, and
Nanoengineering: The Skills and Tools Making Technology Invisible
Copyright ©
Nanowerk LLC
By
Michael
Berger
– Michael is author of three books by the Royal Society of Chemistry:
Nano-Society: Pushing the Boundaries of Technology,
Nanotechnology: The Future is Tiny, and
Nanoengineering: The Skills and Tools Making Technology Invisible
Copyright ©
Nanowerk LLC
|
|||||||
|
Become a Spotlight guest author! Join our large and growing group of guest contributors. Have you just published a scientific paper or have other exciting developments to share with the nanotechnology community? Here is how to publish on nanowerk.com. |
|||||||
