| Jan 15, 2019 | |
A solid-state nanopore platform for digital data storage |
|
| (Nanowerk Spotlight) Demand for data storage is growing exponentially, but the capacity of existing storage media is not keeping up. The digital universe – all digital data worldwide – has grown to over 16 zettabytes (1021 bytes) in 2017. | |
| Most of the world’s data today is stored on magnetic and optical media. Some of the densest forms of storage available commercially are on tape cartridges with about 10GB/mm3. Recent research reported feasibility of optical discs capable of storing 1 petabyte (1015 bytes), yielding a density of about 100GB/mm3. Storing zettabytes of data with this technology would still take millions of units and use significant physical space. | |
| But storage density is only one aspect of archival: durability is also critical. Rotating disks are rated for 3–5 years, and tape is rated for 10–30 years. Current long-term archival storage solutions require refreshes to scrub corrupted data, to replace faulty units, and to refresh technology. To continue preserving the digital universe will require significant advances in storage density and durability. | |
| This is where synthetic DNA comes in. | |
| Using DNA to directly store data is an attractive possibility because it is extremely dense, with a raw limit of 1 exabyte/mm3 (109 GB/mm3), and long-lasting, with observed half-life of over 500 years (read more: "Team stores digital images in DNA – and retrieves them perfectly"). | |
| The write process for DNA storage maps digital data into DNA nucleotide sequences (a nucleotide is the basic building block of DNA), synthesizes the corresponding DNA molecules, and stores them away. Reading the data involves sequencing the DNA molecules and decoding the information back to the original digital data. | |
| Currently, though, synthesizing and sequencing DNA molecules for storing large amounts of data involves complex devices and is very expensive. | |
| In an effort to make data storage in DNA more affordable and commercially viable, researchers at the University of Cambridge have combined nanopore sensing and DNA nanotechnology in a solid-state nanopore platform for digital data storage. This digital data storage method provides an alternative to information storage in the DNA base sequence. | |
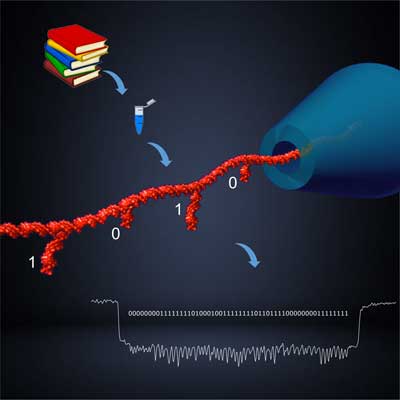 |
|
| Schematic depiction of the team's solid-state nanopore platform for digital data storage. (Image: Kaikai Chen and Alexander Ohmann) | |
| The team, led by Professor Ulrich Keyser at the Cavendish Laboratory, has published their findings in Nano Letters ("Digital Data Storage Using DNA Nanostructures and Solid-State Nanopores"). | |
| "We use high-density DNA hairpins as digital bits representing ‘0’ and ‘1’ along a strand," Kaikai Chen, the paper's first author, explains to Nanowerk. "The strands, that we named DNA carriers, can be built by mixing DNA scaffolds and oligonucleotides to create a library of a large number of different DNA molecules to store information." | |
| "We show that our nanopore has the capability of detecting DNA hairpins down to a length of around 3 nm to decode the digital information," he continues. "The mixing method for DNA structure synthesis allows for convenient data encoding and the nanopore-sensing method directly translates the encoded structure information into electrical signals with great potential for miniature scale integration." | |
| The team points out that their solid-state nanopore platform outperforms previous ones in identifying DNA nanostructures along a DNA strand in terms of resolution and data quality: "We were able to detect DNA hairpins with 8 and 16 bp stems and even distinguish between them on the same molecule." | |
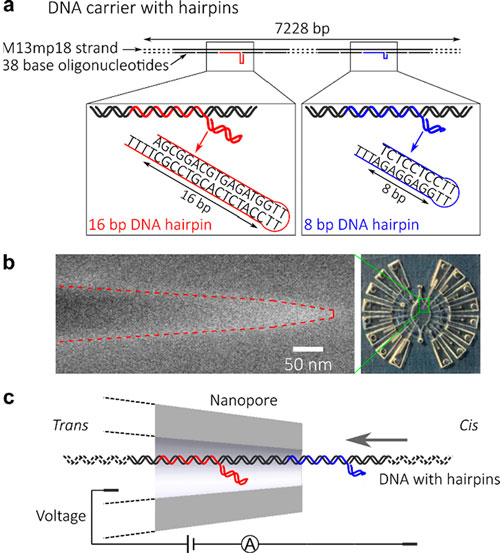 |
|
| Design of DNA carriers with attached DNA hairpins and nanopore measurement. (a) A DNA carrier with DNA hairpins with 16 bp (red) and 8 bp (blue) stems. Structures and sequences of the hairpins are shown at the bottom. (b) The SEM image of a glass nanopore and the integrated chip. Dashed lines indicate the outer edges (scale bar: 50 nm). (c) Schematic of the measurement of the DNA carrier with a nanopore. (Reprinted with permission by American Chemical Society) (click on image to enlarge) | |
| Compared to storing information in the DNA base sequence, this method has advantages due to the ease of encoding and decoding by combining the simple mixing of oligonucleotides with nanopore sensing, which avoids complex devices and the use of enzymes and can be integrated at a miniature scale. | |
| While Keyser's team is working on improving the accuracy of this method by reducing the nanopore decoding errors, they are also looking at DNA computing where the results are read by nanopores. When combined with the DNA storage shown in this work, this could eventually lead to the development of futuristic DNA computers. | |
 By
Michael
Berger
– Michael is author of three books by the Royal Society of Chemistry:
Nano-Society: Pushing the Boundaries of Technology,
Nanotechnology: The Future is Tiny, and
Nanoengineering: The Skills and Tools Making Technology Invisible
Copyright ©
Nanowerk LLC
By
Michael
Berger
– Michael is author of three books by the Royal Society of Chemistry:
Nano-Society: Pushing the Boundaries of Technology,
Nanotechnology: The Future is Tiny, and
Nanoengineering: The Skills and Tools Making Technology Invisible
Copyright ©
Nanowerk LLC
|
|
|
Become a Spotlight guest author! Join our large and growing group of guest contributors. Have you just published a scientific paper or have other exciting developments to share with the nanotechnology community? Here is how to publish on nanowerk.com. |
|
