| Posted: Dec 16, 2008 | |
There is something fishy about these nanotubes |
|
| (Nanowerk Spotlight) Researchers have demonstrated that salmon DNA can be used to develop a simple and scalable method for sorting carbon nanotubes that reduces the cost, as compared to commonly used synthetic DNA, by a factor of 1,000. | |
| Before carbon nanotubes (CNTs), especially single-walled ones, can live up to the many expectations for their use in nanoelectronics, researchers have to overcome a seemingly trivial but nonetheless major obstacle: how to separate a produced batch of nanotubes according to their properties such as diameter, length, chirality and electronic attributes. Current production methods for CNTs result in a jumble of units with different properties, all lumped together in bundles, and often blended with some amount of amorphous carbon. These mixtures are of little practical use since many advanced applications, especially for nanoelectronics, are sensitively dependent on tube structures and the slightest deviation from a desired set of parameters can lead to vastly different performance results. | |
| Accomplishing a purification of 99.999% requires a highly effective surfactant system or nanotube chemical functionalization. In a previous Spotlight we have introduced you to techniques that use DNA to sort carbon nanotubes. Scientists have developed a technique where oligonucleotide sequences self-assembles into a helical structure around individual nanotubes, creating DNA-CNT hybrids with electrostatic properties that depend upon the tube diameter and electrical properties. CNTs can be separated on the basis of these properties using anion exchange chromatography. The separation of metallic and semiconducting nanotubes is improved compared with other techniques, and separation on the basis of tube diameter has become possible. | |
| "The price for the single-stranded d(GT)20 DNA oligomer, which is an effective separation agent, is typically $25,000 per gram and usually oligo-DNA assisted SWCNT dispersion experiments are carried out with a DNA:SWCNT weight ratio of 1:1, discarding the majority of unbound DNA," Dr. Rajesh R. Naik tells Nanowerk. "This poses an extremely high price of $25,000 for oligo-DNA in treating every gram of carbon nanotubes, and thus, a cost-effective nucleic acid system is highly in demand." | |
| Naik, a scientist at the Air Force Research Laboratory (AFRL) at Wright-Patterson Air Force Base in Dayton, Ohio, together with his team from the Materials and Manufacturing Directorate, has described a rather simply, scalable technique that can be used in the enrichment of carbon nanotubes using genomic DNA. They have published their work in the November 19, 2008 online issue of Nano Letters ("A Enrichment of (6,5) Single Wall Carbon Nanotubes Using Genomic DNA"). | |
 |
|
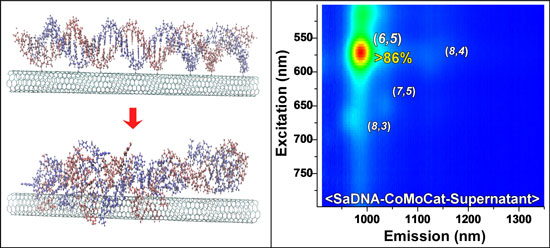
| The image on the left shows a model of the DNA interacting with the SWCNTs using classical all-atom molecular dynamics simulations and the right panel is the normalized photoluminescence excitation emission contour plot of DNA d(GT)20 dispersed SWCNTs (Image: Dr. Naik, AFRL) | |
| In contrast to previously published work, that described the use of nucleic acids to enrich certain classes of SWCNTs, Naik's team uses genomic DNA as the starting material as opposed to designed oligonucleotides. | |
| "Based on our findings, where we demonstrate that the genomic DNA exfoliates and disperses SWCNTs on a level comparable to d(GT)20 oligomer DNA, it appears that the chemical composition (GC content) of the DNA is critical in selecting certain classes of carbon nanotubes," says Naik. "Equally important is the size of the DNA, since this also affects chirality separation." | |
| The biggest impact, though, comes from the lowered cost of using genomic DNA as compared to synthetic DNA. Rather than having to spend $25,000 on synthetic designer oligonucleotides, the team conducted their solubilization and separation study by using a genomic salmon DNA, which is a byproduct of the fishing industry and costs about $20 per gram. | |
| Naik and his colleagues at the AFRL have been working on making devices out of DNA films containing SWCNTs of a given chirality. Confronted with the challenges of purifying their samples, they decided to develop a new strategy for sorting SWCNTs that is scalable, non destructible, and economical at the same time. Having accomplished that, the researchers now want to find out if they can fabricate devices using SWCNTs of a given chirality and diameter from their DNA-separated samples. | |
| Another aspect of their future research work will be to find out what DNA sequences and molecular weight preferences can be uncovered using this method. Interestingly, the particular genomic DNA that the scientists used specifically enriches only (6,5) tubes but not tubes of similar diameter. Therefore, understanding the enrichment mechanism and whether one can rationally design DNA sequences using computational tools, will be important research goals. | |
| "Clearly, the GC content of the DNA has an important role to play in SWCNT separation, but just having a defined GC content might not be sufficient for chirality enrichment" says Naik. "DNA fragment length and sequence composition could also be important parameters to consider." | |
 By
Michael
Berger
– Michael is author of three books by the Royal Society of Chemistry:
Nano-Society: Pushing the Boundaries of Technology,
Nanotechnology: The Future is Tiny, and
Nanoengineering: The Skills and Tools Making Technology Invisible
Copyright ©
Nanowerk LLC
By
Michael
Berger
– Michael is author of three books by the Royal Society of Chemistry:
Nano-Society: Pushing the Boundaries of Technology,
Nanotechnology: The Future is Tiny, and
Nanoengineering: The Skills and Tools Making Technology Invisible
Copyright ©
Nanowerk LLC
|
|
Become a Spotlight guest author! Join our large and growing group of guest contributors. Have you just published a scientific paper or have other exciting developments to share with the nanotechnology community? Here is how to publish on nanowerk.com.
