| Posted: Jan 29, 2009 | |
DNA nanotechnology goes 3D |
|
| (Nanowerk Spotlight) DNA, the fundamental building block of life, has become an intense nanotechnology research field. DNA molecules can serve as precisely controllable and programmable scaffolds for organizing functional nanomaterials in the design, fabrication, and characterization of nanoscale devices such as sensors and electronics (see: "Nanotechnology cut and paste with single molecules" or "DNA electronics in nanotechnology"). | |
| Most DNA research on controlled self-assembly deals with two-dimensional, i.e. flat, patterns and an expansion of these arrays into the third dimension has been challenging. New research coming out of UC Santa Barbara describes the self-assembly of multilayer hexagonal DNA arrays through highly regular interlayer packing. | |
| "We found that DNA arrays assembled into a two dimensional hexagonal pattern, or a sheet, assemble further into multilayer stacks," Norbert Reich tells Nanowerk. "What is more interesting is that these multilayer stacks have certain rules for assembly. The DNA sheets interact with one another at specific rotational and translational orientations." | |
| Unlike the formation of DNA crystals or DNA hydrogels, layer by layer assembly has an advantage as various layers can pack within three-dimensional structures, thereby providing different pore sizes for guest molecule incorporation. | |
| Reporting their work in Langmuir ("Self-Assembly of DNA Arrays into Multilayer Stacks"), Reich and his team point out that high resolution visualization of these structures is beyond the capability of atomic force microscopy, but they believe three-dimensional architectures to be one of the exciting directions of the nucleic acid nanotechnology. | |
| "Artificial nucleic acid architectures have been mostly restricted to a plane" explains Alexey Koyfman, first author of the paper (he was a graduate student in Reich's group and is now a postdoctoral fellow at Baylor College of Medicine in Houston, Texas). "Most of these structures are imaged with atomic force microscopy (AFM) but 3D objects are not easily visualized by AFM. Recently, 3D objects have been assembled as whole objects and imaged by cryo electron microscopy. Our findings now demonstrate that it is possible to make 3D architectures using layer by layer assembly of DNA sheets." | |
| The team was successful at visualizing and identifying three layers but they mention that they have also seen assemblies composed of more than three sheets. | |
 |
|
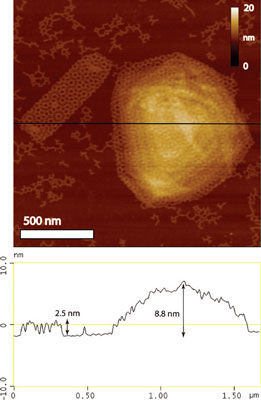
| AFM and cross section of two-layer and multilayer assemblies. Multilayer assemblies form at low Mg2+ concentration. The cross section (black line) was shown to represent the height of a two-layer assembly (2.5 nm) rotated 30° with respect to one another and a taller multilayer assembly that is 8.8 nm in height. (Reprinted with permission from American Chemical Society) | |
| "AFM was not a good tool to accurately show how many layers these architectures had" says Koyfman. "Hopefully techniques such as cryo electron tomography can help in the visualization of multilayer architectures." | |
| One of the challenges in the field is to assemble nucleic acids into large uniform arrays. The structures described by Reich's team are made of a 3-point star DNA motif that could extend microns across with the same repeating hexagonal pattern of a honeycomb pattern. | |
| "We have previously reported that positive gold nanoparticles can fit within negative nucleic acid architectures" says Reich ("Controlled Spacing of Cationic Gold Nanoparticles by Nanocrown RNA"). "This work concentrated on 5 nm particles that can fit within 5 nm nucleic acid architectures. We wanted to try the larger hexagonal DNA architecture to fit larger nanoparticles using electrostatic interactions. So we were aiming to make hexagonally positioned nanoparticles in a plane that could extend microns across." | |
| In their experiments, the scientists found that slow cooling of a mixture of three single-stranded DNA sequences in the presence of Mg2+ results in formation of 3-point star motifs that self-assemble into 2D hexagonal DNA arrays which furthermore form diverse multilayer architectures. | |
| "The size of the two-dimensional arrays and subsequent stacking to form multilayer structures are highly dependent on Mg2+ concentration" explains Koyfman. "We found that DNA bilayers and multilayers of defined shape are favored in 2-5 mM Mg2+ with an average lateral size of 700 nm. Arrays are much larger, up to 20 µm across, in 10-15 mM Mg2+." | |
| There are a number of potential applications for this kind of research. One is the organization of inorganic materials on the DNA architecture, i.e. using DNA as scaffolds. Due to the multilayer packing of DNA sheets it is theoretically possible to position materials within the three dimensional framework of the DNA. | |
| Koyfman mentions that it would also be interesting to investigate the bulk properties of DNA or slightly modified nucleic acids. "Nucleic acids carry information and take advantage of hydrogen bonds to form a double helix. Nucleic acids could prove to be macroscale functional materials due to their nanoscale properties." | |
| Reich's group has also been working on DNA nanotechnology by attaching DNA structures to cancer cells. The attached DNA array could possibly carry a number of therapeutic molecules that could then penetrate a tumor cell. The way Koyfman puts it, "the cell in a way has a DNA 'band-aid' with potential cargo attachment sites". They are in the process of submitting this paper, so details about this work are not available yet. | |
 By
Michael
Berger
– Michael is author of three books by the Royal Society of Chemistry:
Nano-Society: Pushing the Boundaries of Technology,
Nanotechnology: The Future is Tiny, and
Nanoengineering: The Skills and Tools Making Technology Invisible
Copyright ©
Nanowerk LLC
By
Michael
Berger
– Michael is author of three books by the Royal Society of Chemistry:
Nano-Society: Pushing the Boundaries of Technology,
Nanotechnology: The Future is Tiny, and
Nanoengineering: The Skills and Tools Making Technology Invisible
Copyright ©
Nanowerk LLC
|
|
|
Become a Spotlight guest author! Join our large and growing group of guest contributors. Have you just published a scientific paper or have other exciting developments to share with the nanotechnology community? Here is how to publish on nanowerk.com. |
|
