| Aug 12, 2019 |
Bacteria made to mimic cells, form communities
|
|
(Nanowerk News) Rice University scientists have found a way to engineer a new kind of cell differentiation in bacteria, inspired by a naturally occurring process in stem cells.
|
|
They have created a genetic circuit able to produce genetically distinguished cells of Escherichia coli as the bacterium divides. By controlling this process, it is possible to create diverse communities of microbes that exhibit complex, non-native behaviors.
|
|
Rice synthetic biologist Matthew Bennett and Sara Molinari, a former student in the university’s Systems, Synthetic and Physical Biology Ph.D. program, led the project to show how manipulating the genetic code of plasmids — free-floating pieces of circular DNA in cells — can be used to obtain stem cell-like differentiation in bacteria.
|
|
“Stem cells have the remarkable ability to divide asymmetrically,” Bennett said. “Upon division, the original stem cell stays the same, but the new daughter cell has a completely new phenotype. That’s asymmetric cell division, and multicellular organisms use it to help control their cellular makeup.
|
|
“As a synthetic biologist, I think a lot about creating and controlling differentiated cell types within a multicellular population,” he said. “Here, we’ve taken what we know about stem cells and engineered the means to do it in bacteria.”
|
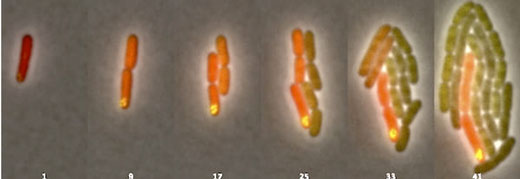 |
| Over the course of 93 minutes, the process Rice University researchers call asymmetric plasmid partitioning prompted a single Escherichia coli bacterium to divide into two genetically distinct types of bacteria. Daughter microbes seen fluorescing in the right images retain the DNA-carrying plasmids (marked by the yellow dots) while their now-differentiated siblings do not. (Image: The Bennett Lab)
|
|
The researchers reported the development, which they call asymmetric plasmid partitioning (APP), in Nature Chemical Biology ("A synthetic system for asymmetric cell division in Escherichia coli").
|
|
Molinari first discovered how to force plasmids in E. coli to aggregate in a single cluster so they do not distribute homogeneously during cell division, but rather are inherited by only one of the two daughter cells. The plasmid-laden daughter cell remains identical to the progenitor cell, while its sibling becomes genetically distinct as it loses the genetic information present on the plasmids.
|
|
She then expanded the synthetic circuit to induce the simultaneous asymmetric partitioning of two plasmid species in a single cell, resulting in four genetically distinct E. coli. Some of the cells have motility programmed in; they can literally go their own way and help form patterns in the resulting colony.
|
|
“When we started, we were thinking about creating materials that have to be able to sense and adapt to an environment,” said Molinari, who recently earned her doctorate at Rice. “We thought if we could mimic this feature of higher-order tissues, we would increase the robustness of our colonies and their ability to perform tasks. The challenge was to engineer a population of bacteria that becomes something else whenever it’s needed.”
|
|
Molinari and her colleagues hit the jackpot on their first try with E. coli. “There was no canonical way to engineer asymmetrical cell division,” she said. “It was a crazy idea, and it magically worked the first time.
|
|
“But there was something we couldn’t completely figure out about the system,” Molinari said. “It took two years to find out I made a cloning mistake when I got this protein and put it in my plasmid. I had randomly added 17 amino acids at the beginning of the protein, and that made the whole system work.”
|
|
With that knowledge, she proceeded to improve upon the hydrophobic proteins that cluster in cells while they bind to target plasmids, holding them in place.
|
|
Bennett noted natural processes either load enough plasmids into a cell to ensure some land in each daughter cell or actively pull plasmids into each of the new cells to ensure they remain identical. “We have shown we can outcompete those processes,” he said.
|
|
APP could turn simple organisms into complicated systems that enhance understanding of multicellular life. “We’re pretty good at designing bacteria,” Bennett said. “We’ve been doing that for years now. I think the field has evolved to the point where we can do amazing things with bacteria and people are asking what else we can do.”
|
|
The new discovery, he said, provides a path forward.
|
|
“There are three main hallmarks to multicellular life,” he said. “One is differentiation through asymmetric cell division. Another is intercellular communication, which synthetic biologists have been engineering for years. And the third is cell adhesion, so cells stay where they’re supposed to and stick to each other. If we can control all those things together, we can talk about engineering interesting multicellular lifeforms.
|
|
“It starts to feel a bit like science fiction, for sure,” he said.
|

