| Aug 19, 2019 |
Microorganisms build the best fuel efficient hydrogen cells
|
|
(Nanowerk News) For all the advances technology has made throughout our lives, in many cases it stands behind what nature can do. Ants can carry 5000 times their weight, and spider webs are five times stronger than steel. Fuel efficiency is no different.
|
|
In a new study seen in Angewandte Chemie International Edition ("Cysteine SH and Glutamate COOH Contributions to [NiFe] Hydrogenase Proton Transfer Revealed by Highly Sensitive FTIR Spectroscopy"), researchers from Nara Institute of Science and Technology (NAIST) report new details on the proton transfer pathway of nickel-iron [NiFe]-hydrogenase using Fourier transfer infrared spectroscopy (FT-IR). This transfer is crucial for the hydrogen metabolism of microorganisms, and the study gives scientists a better understanding of how to mimic nature in the construction of new biofuel cells.
|
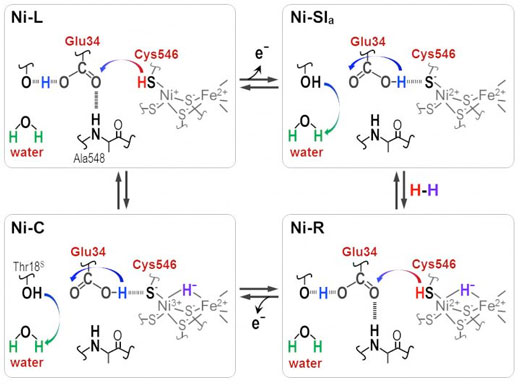 |
| Protons are transferred between the NiFe active site and protein surrounding through the Cyt546 and Glu34 residues during the catalytic reaction of [NiFe] hydrogenase. (Image: Shun Hirota)
|
|
Hydrogen metabolism is one of the oldest forms of energy production for life and one of the most intensely studied because of its significance in evolution. It has also attracted attention as it is a clean energy source. Among the enzymes responsible for hydrogen metabolism, [NiFe]-hydrogenase is the most abundant and ancient.
|
|
Much is known about the enzyme. It reversibly oxidases hydrogen atoms through its Ni-Fe active site, which is surrounded by specific amino acids. However, notes NAIST Professor Shun Hirota, who managed the study, certain fundamentals in the chemical reactions need to be understood before using this nanomachine to design biofuel technologies.
|
|
"Hydride and proton coordinate with the Ni-Fe site in the protonation-deprotonation cycle, resulting in four hydrogenase states. But the proton transfer pathway remains unknown," he says.
|
|
One reason for the lack of clarity has been an inability to resolve stretching frequencies of different hydrogen bonds. In the new work, Hirota's team, in collaboration with other researchers in Japan and China, solved this problem by observing photoconversions of three hydrogenase states with FT-IR spectra.
|
|
Distinct absorbance frequencies revealed that one of the four cysteines, cysteine 546, along with another amino acid, glutamic acid 34, and an ordered water molecule are crucial for the transfer. The scientists used the new information to deduce how the sulfur molecule of cysteine 546 and the acid group in glutamic acid 34 form hydrogen bonds to regulate the hydrogen transport.
|
|
"Our results show that cysteine 546 is a proton donor and acceptor in the [Ni-Fe] hydrogenase cycle. They also demonstrate how a low-barrier hydrogen bond for glutamic acid 34 is formed and cleaved during the catalytic cycle," notes Hirota.
|
|
Microorganisms have adapted to a wider range of environmental conditions than humans. They can be found in the comfortable climate of our bedrooms to the most hostile environments on earth. Thus, seeing how the [Ni-Fe] site of [Ni-Fe] hydrogenase transfers protons optimally provides a paradigm for researchers seeking to manufacture high efficiency energy catalysts under a host of conditions.
|
|
"Life has spent billions of years adapting to energy. We believe it gives us the best model for energy efficiency in the future," says Hirota.
|

