| Posted: Apr 20, 2010 | |
Atomistic model of spider silk nanostructure paves way for new class of synthetic materials |
|
| (Nanowerk Spotlight) Material scientists have been fascinated by spider silks for a long time – ultra-strong and extensible self-assembling biopolymers that outperform the mechanical characteristics of many synthetic materials, including steel. While the source of these unique material properties are thought to lie in the distinct protein structures found in spider silk (read more: "Protein engineering - from the humble spider to the nanotechnology future of material design"), the physical mechanisms behind them have remained poorly understood for decades. This is partly due to the fact that structural models of spider silk with atomic-level resolution have not been available, preventing a molecular-level analysis. | |
| So far, only small models of isolated crystalline domains of silk have been reported. Yet, the integrated structure of the semi-amorphous regions combined with crystalline domains in silk is critical for properties such as toughness and fracture mechanisms. To unravel silk's secrets and ultimately be able to create synthetic materials that duplicate, or even exceed, the extraordinary properties of natural silk, researchers need to fully understand the links between genetic makeup, chemical interactions, and structure, as well as its macroscale mechanical properties. | |
| Setting the stage for full-atomistic studies on silk domains that will contribute toward an improved understanding of the source of the strength and toughness of this amazing biological material, MIT researchers Sinan Keten and Markus J. Buehler have developed and applied a framework for predicting the nanostructure of spider silk using atomistic principles. | |
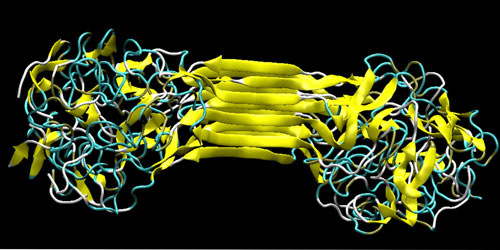 |
|
| Molecular structure of silks, showing the characteristic beta-sheet nanocrystal (yellowish) and the semi-amorphous domains. The figure depicts the self-assembled structure of several strands of MaSp1 protein, a result obtained from replica exchange molecular dynamics simulation. (Image: Markus J. Buehler, MIT) | |
| Buehler, the Esther and Harold E. Edgerton Associate Professor in MIT’s Department of Civil and Environmental Engineering, explains to Nanowerk that these findings are widely applicable not only to different types of silks but could also to other complex heterostructured biopolymers. | |
| In a paper published as the cover article in Applied Physics Letters on April 12, 2010 ("Atomistic model of the spider silk nanostructure"), Keten and Buehler demonstrate an innovative application of replica exchange molecular dynamics simulations on a key spider silk repeating sequence, resulting in the first atomistic level structure of spider silk. This represents an important breakthrough in the elucidation of the spider silk nanostructure, based on experimentally validated bottom-up atomistic simulation. This advance was made possible due to the availability of ultra large-scale computational resources, which now permit the identification of the molecular structure of complex protein materials such as silk. | |
| Buehler explains that In most engineered materials – ceramics, for instance – high strength comes with brittleness. "Once ductility is introduced, materials become weak" he says. "But not silk, which has high strength despite being built from inherently weak building blocks. It turns out that’s because these building blocks – the tiny beta-sheet crystals, as well as filaments that join them – are arranged in a structure that resembles a tall stack of pancakes, but with the crystal structures within each pancake alternating in their orientation. This particular geometry of tiny silk nanocrystals allows hydrogen bonds to work cooperatively, reinforcing adjacent chains against external forces, which leads to the outstanding extensibility and strength of spider silk." | |
| More specifically, the MIT researchers found the formation of beta-sheet structures in poly-Ala rich parts of the structure, the presence of semi-extended GGX domains that form H-bonded 31 helix type structures and a complete lack of alpha-helical conformations in the molecular structures formed by the self-assembly of MaSp1 proteins. These results resolve controversies around the structure of the amorphous domains in silk, by illustrating for the first time that these semi-extended, well-oriented and more sparsely H-bonded structures that resemble 31 helices could be the molecular source of the large semi-crystalline fraction of silks and the so-called 'pre-stretched' configuration proposed for these domains. | |
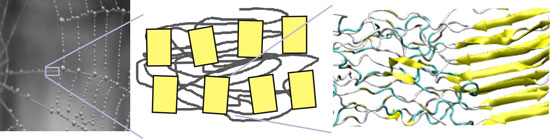 |
|
| Hierarchical structure of spider silk, showing structural features from the molecular to the overall structural scales. The makeup of this material exemplifies the merger of structure and material, a concept universal to most biological materials. (Image: Markus J. Buehler, MIT) | |
| The results allow the mapping of earlier experimental findings to representative structures with atomistic detail, enabling now to probe the fundamental design principles underlying the structural make-up of silk and its link to mechanical properties. | |
| This model resolves the challenge of linking molecular chemistry and genetic sequence of spider silk proteins to atomic resolution structures, paving the way for mechanical characterization of spider silk fibrils. | |
| Most importantly, the structures reported in this recent work will now enable researchers to carry out further characterization using atomistic and molecular simulation techniques. The framework presented in their work could also be applied to other macromolecular structures and as such have broader impact. | |
| "Linking genetic-level information to material properties has remained a major challenge in the field of biological materials, with important implications not only for structural extraorganismal materials such as silk, but also for our understanding of genetic diseases and other disorders", says Buehler. | |
| Buehler says the work has implications far beyond just understanding silk. He notes that the findings could be applied to a broader class of biological materials, such as wood or plant fibers, and bio-inspired materials, such as novel fibers, yarns and fabrics or tissue replacement materials, to produce a variety of useful materials out of simple, commonplace elements. | |
| "We are looking at the possibility of synthesizing materials that have a similar structure to silk, but using molecules that have inherently greater strength, such as carbon nanotubes," says Buehler. | |
| According to him, the long-term impact of this research will be the development of a new material design paradigm that enables the creation of highly functional materials out of abundant, inexpensive materials. This would be a departure from the current approach, where strong bonds, expensive constituents, and energy intensive processing – at high temperatures – are used to obtain high-performance materials. | |
| This work was supported by the Office of Naval Research, with additional funding from the National Science Foundation, the Army Research Office, and the MIT Energy Initiative. The research was supported by an allocation of advanced computing resources supported by the National Science Foundation. | |
 By
Michael
Berger
– Michael is author of three books by the Royal Society of Chemistry:
Nano-Society: Pushing the Boundaries of Technology,
Nanotechnology: The Future is Tiny, and
Nanoengineering: The Skills and Tools Making Technology Invisible
Copyright ©
Nanowerk LLC
By
Michael
Berger
– Michael is author of three books by the Royal Society of Chemistry:
Nano-Society: Pushing the Boundaries of Technology,
Nanotechnology: The Future is Tiny, and
Nanoengineering: The Skills and Tools Making Technology Invisible
Copyright ©
Nanowerk LLC
|
|
|
Become a Spotlight guest author! Join our large and growing group of guest contributors. Have you just published a scientific paper or have other exciting developments to share with the nanotechnology community? Here is how to publish on nanowerk.com. |
|
