| Posted: Mar 12, 2012 | |
DNA reference tags allow single-molecule research on complex genomes |
|
| (Nanowerk Spotlight) Knowing the distribution of DNA binding proteins along the genome is very informative and can tell scientists about the state of gene expression at the time of measurement. These DNA-binding proteins include transcription factors which modulate the process of transcription, various polymerases, nucleases which cleave DNA molecules, and histones which are involved in chromosome packaging in the cell nucleus. | |
| In a previous Nanowerk Spotlight ("Quantum dots light up individual DNA binding proteins") we reported on work that demonstrated the viability of a single-molecule approach to directly visualize and map protein binding sites on DNA using fluorescent quantum dots, allowing multicolor, nanometer-resolution localization. | |
| Yuval Ebenstein, who led this earlier work as a post doctoral fellow at the Department of Chemistry and Biochemistry at UCLA, has now set up his own research group at Tel Aviv University. Together with his collaborators from UCLA and RWTH Aachen University he has recently shown that proteins bound to DNA can be located very accurately by direct imaging. | |
| Reporting their findings in the February 16, 2012 online edition of Angewandte Chemie International Edition ("Enzymatically Incorporated Genomic Tags for Optical Mapping of DNA-Binding Proteins"), the precision of the measurement presents new opportunities for contextual genomic research on the single-molecule level. | |
| "As a single molecule approach we hope to be able to detect very rare genomic configurations that are usually hidden in the noise of conventional bulk methods for mapping DNA binding proteins," Ebenstein tells Nanowerk. "This could have impact in diagnostics and personalized medicine." | |
  |
|
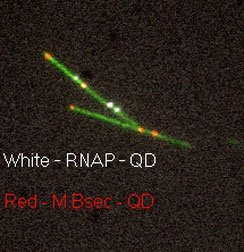
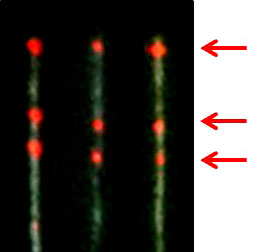
| The images show T7 genomes with the pattern created by quantum dots reference tags in red and Bound T7-RNAP showing in white (Images: Yuval Ebenstein) | |
| Previously, Ebenstein and his collaborators mapped proteins by measuring their distance to the ends of the DNA molecule. | |
| "This meant we were only able to work with small genomes like the genomes of viruses that could be imaged intact" he explains. "In this new work, beyond the fact the precision was improved five-fold relative to previous reports due to the reference tags, we are now not dependent on the ends of the DNA for mapping and can work on more complex genomes such as the human genome which is over one and a half meters long." | |
| In order to do this, the scientists use an enzyme to attach a biotin to specific sequences in a viral genome. They then send in quantum dots that attach to the biotin and create a fluorescence "barcode" along the genome. They use this pattern to accurately map the positions of individual RNA polymerases sitting on the genome and labeled with quantum dots of a different color. | |
| "A very cool aspect of this work is that the reference tags were incorporated into the DNA by enzymes that were tricked into attaching a biotin to the DNA at specific locations," says Ebenstein. "This is a great demonstration of how combining chemistry, biology and physics can yield new applications by tweaking natural systems like enzymatic molecular motors and harnessing their ability to manipulate matter at the nanoscale." | |
| For their work, the team used the T7 bacteriophage genome as a model system to demonstrate the utility of this approach to the assignment of T7 individual RNA polymerase (RNAP) binding sites. The objective was to generate a unique fluorescent pattern along the T7 genome that would be sparse enough to not produce overlapping fluorescence signals and would also not interfere with RNAP binding. | |
| In order to be useful, this method has to be able to collect and analyze huge amounts of data in a short time. The current approach relies on stretching the DNA molecules on a glass surface and imaging them on a fluorescence microscope and is relatively slow. | |
| The researchers are therefore working on a new high-throughput approach where DNA is squeezed through thousands of parallel nanochannels which cause it to straighten and create the optical barcode "on the fly" while still suspended in the channel. This way, new material can constantly flow through the channels and very large data sets may be acquired. | |
 By
Michael
Berger
– Michael is author of three books by the Royal Society of Chemistry:
Nano-Society: Pushing the Boundaries of Technology,
Nanotechnology: The Future is Tiny, and
Nanoengineering: The Skills and Tools Making Technology Invisible
Copyright ©
Nanowerk LLC
By
Michael
Berger
– Michael is author of three books by the Royal Society of Chemistry:
Nano-Society: Pushing the Boundaries of Technology,
Nanotechnology: The Future is Tiny, and
Nanoengineering: The Skills and Tools Making Technology Invisible
Copyright ©
Nanowerk LLC
|
|
|
Become a Spotlight guest author! Join our large and growing group of guest contributors. Have you just published a scientific paper or have other exciting developments to share with the nanotechnology community? Here is how to publish on nanowerk.com. |
|
