| Posted: May 09, 2013 | |
Single-molecule nanopore research made easy |
|
| (Nanowerk Spotlight) Nanopores are an exciting class of single molecule nanosensors. For several years now, nanopore technology has been developed as a biosensor at the single-molecule resolution to detect an array of biomedical molecules, such as DNA, RNA, protein, bio-toxin (see for instance "For the first time single proteins were detected with nanopores" or "Novel, fast DNA nanopore detector features integrated tunneling electrodes"), and various nanopore projects have been funded to develop the next generation of DNA sequencing technology. | |
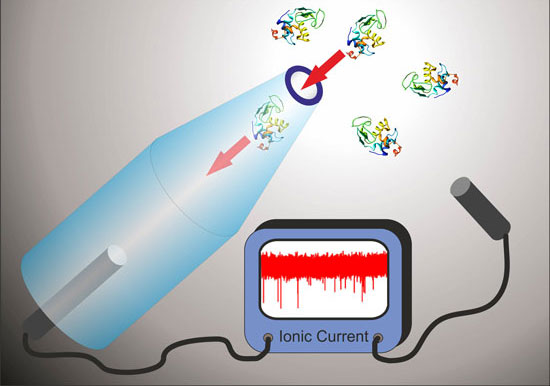
| The sensing principle is based on the resistive pulse technique – molecules are detected as they pass through a single nanopore since during translocation the molecules exclude ions and therefore modulate the current. | |
| In new work, researchers in the UK have demonstrated the single molecule detection of a wide range of proteins with solid state glass nanopores. Solid state nanopores are more applicable to protein analysis than biological nanopores as these are normally limited to diameters of <2 nm. | |
| So far, the main complication in nanopore research has been the supply of the nanopores. This new work now demonstrates that protein analysis – on the single molecule level and label-free – can be performed by anybody with a pipette puller and a decent electrophysiology amplifier. | |
| The detection, analysis and identification of proteins is a long-standing issue in the life sciences in general. The standard approaches use large quantities of proteins or need complicated labelling techniques. With the new method reported here, these challenges are overcome. The proteins can be analyzed in the native conformation, if necessary in close to physiological conditions without any chemical modification. | |
| Reporting their work in the April 22, 2013, online edition of ACS Nano ("Single Protein Molecule Detection by Glass Nanopores"), a team led by Ulrich Keyser, a University Lecturer at the Cavendish Laboratory at the University of Cambridge, demonstrate single molecule detection of proteins. These experiments extend the range of ionic current detection of glass nanopores, which are simple and inexpensive alternatives to those formed by ion or electron beam drilling in silicon nitride membranes. | |
 |
|
| Glass nanopores can be used to detect and analyze single molecules in solution. (Image: Dr. Keyser, University of Cambridge) | |
| Keyser and his team fabricated glass nanopores by laser-assisted capillary pulling and showed that they can be produced with average inner diameters of just 17 nm. | |
| "This is a remarkably simple table-top technology for interfacing with the nanoworld since no clean room facilities or electron beam equipment is needed," Keyser tells Nanowerk. "It is also cheap with each capillary costing about $1 and the fabrication is high throughput since the laser pulling process takes just under one second. This contrasts with traditional solid state nanopores made in silicon based membranes which require transmission electron microscopy for drilling nanopores." | |
| Recent work in Keyser's group demonstrated that glass nanopores can be integrated into a 16 channel device – the first published demonstration of multiplexed single molecule ionic current sensing with solid state nanopores (see "Multiplexed ionic current sensing with glass nanopores"). In this work the team also showed that they could trap DNA origami nanostructures onto glass nanopore tips to form hybrid nanopore architectures (see: "DNA Origami Nanopores"). | |
| Not only does the new technique show that a simple table-top technology like glass nanopores can be applied to the measurement of a wide range of protein molecules; it also provides a greater understanding of the translocation of proteins through nanopores. | |
| "We show that many protein translocations are not detected due to the fast translocation dynamics," says Keyser. "Interestingly, in our experimental conditions, electrophoresis dominates over electroosmotic effects in determining the direction of translocation of the proteins. Finally we demonstrate the detection of the protein PrP, which in its abnormal form is associated with transmissible spongiform encephalopathies, such as scrapie in sheep, bovine spongiform encephalopathy (BSE) in cattle, and Creutzfeldt?Jacob disease (CJD) of humans. This provides a basis for nanopore measurements of the aggregation of PrP." | |
| This work can be applied as a simple label free technique for protein and amyloid characterisation. It also paves the way for single molecule studies of protein aggregation. | |
| "The study of protein aggregation with nanopores is a potential future direction for the field," Keyser points out. "Nanopores have great potential as a label-free, in-solution, high-throughput and single-molecule technique which can reveal the kinetics of protein aggregation. This is highly relevant for understanding a number of diseases related to protein aggregation such as Alzheimers and BSE." | |
| He also notes that challenges include control of the surface chemistry of the nanopore to slow the passage of protein molecules through the nanopore. Also controlled attachment of binding sites that can capture specific protein molecules would be highly desirable so that individual proteins can be measured from a mixture. | |
 By
Michael
Berger
– Michael is author of three books by the Royal Society of Chemistry:
Nano-Society: Pushing the Boundaries of Technology,
Nanotechnology: The Future is Tiny, and
Nanoengineering: The Skills and Tools Making Technology Invisible
Copyright ©
Nanowerk LLC
By
Michael
Berger
– Michael is author of three books by the Royal Society of Chemistry:
Nano-Society: Pushing the Boundaries of Technology,
Nanotechnology: The Future is Tiny, and
Nanoengineering: The Skills and Tools Making Technology Invisible
Copyright ©
Nanowerk LLC
|
|
|
Become a Spotlight guest author! Join our large and growing group of guest contributors. Have you just published a scientific paper or have other exciting developments to share with the nanotechnology community? Here is how to publish on nanowerk.com. |
|
