| Posted: Nov 02, 2012 | |
Proteins interact with 'ultrasmall' nanomaterials in unique ways |
|
| (Nanowerk Spotlight) Proteins are one of the fundamental functional components of a biological system. They catalyze chemical reactions, participate in signaling between cells, and perform mechanical work. Most biological environments contain a diverse collection of proteins of varying concentration, composition, structure, and function. | |
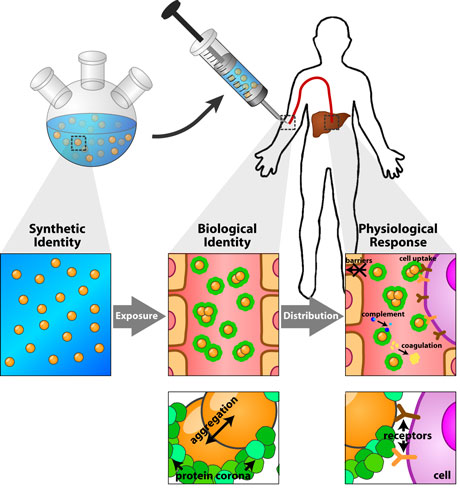
| When a nanomaterial enters a biological environment, it comes into immediate contact with a collection of proteins that is characteristic of that environment. If the nanomaterial possesses high surface energy, by being charged or hydrophobic for example, it will interact with a number of these proteins. Most protein-nanomaterial interactions lead to some alteration in protein structure, known as a conformational change. Conformational changes can activate or modulate enzymatic cascades and signaling pathways. They can also alter protein function or induce protein aggregation. The outcome of protein-nanomaterial interactions is an important determinant of the biological response to a nanomaterial (see Fig. 1). | |
 |
|
| Figure 1: Schematic depicting the formation of the protein corona around a nanoparticle and its effect on the subsequent biological response. Image reprinted from "Understanding and controlling the interaction of nanomaterials with proteins in a physiological environment " with permission from Royal Society of Chemistry. | |
| A protein may become associated with the nanomaterial surface during a protein-nanomaterial interaction, in a process called ‘adsorption’. The set of proteins adsorbed to the surface of a nanomaterial at any given time is known as the protein ‘corona’. The protein corona is conceptually divided into ‘hard’ and ‘soft’ components. The hard component refers to proteins that remain tightly-associated with the nanomaterial surface, returning slowly to the surrounding environment. The soft component refers to loosely-adhered proteins that exchange rapidly with the surrounding environment. The hard corona defines, in part or in whole, the interface between a nanomaterial and its surrounding biological environment, and changes the way in which the nanomaterial interacts with biomolecules and cells within that environment. | |
| Depending on its composition, the protein corona can influence the biological response to a nanomaterial in different ways. The protein corona can have a beneficial effect if it enhances the interaction of a nanomaterial with a target cell type or biomolecule, or suppresses the interaction of a nanomaterial with an off-target cell type or biomolecule (read more in this Nanowerk Spotlight: "The cellular response to nanomaterials depends on the environment"). However, the protein corona can also have a detrimental effect if it mediates undesired cellular interactions, protein aggregation, loss of protein function, or aberrant signaling. Characterizing the composition and structure of the protein corona, and its dependence on the physical and chemical properties of a nanomaterial is vital to understanding and controlling the interaction of nanomaterials with biological systems in the human body and the environment. | |
| For a given biological environment (such as blood, interstitial fluid, or cellular cytoplasm), the composition of the protein corona depends on the chemical composition and structure of the nanomaterial surface. The identities and quantities of the proteins present in the protein corona, along with their conformation, arrangement, structure, and orientation can be used to predict which cell types and biomolecules a nanomaterial is likely to interact with, along with the outcome of these interactions. By relating the properties of the protein corona to the chemical characteristics of a nanomaterial, principles can be developed to guide the design of nanomaterials with controlled protein interactions and biological behavior. | |
| When researchers perform an experiment to characterize the composition of the protein corona, they typically mix a nanomaterial with a biofluid, incubate the two for long enough to allow protein adsorption, and then purify nanomaterial-protein complexes from unbound proteins. Once the nanomaterial-protein complexes are isolated, the proteins can be stripped from the nanomaterial surface and identified and/or quantified using proteomics techniques such as Western Blot or shotgun mass spectrometry. | |
| The separation of protein-nanomaterial complexes from unbound proteins is a critical step in the characterization process. If the nanomaterial-protein complexes cannot be efficiently purified from unbound proteins, the subsequent analysis will characterize nanomaterial-bound proteins as well as proteins that are not associated with the nanomaterial. Unbound proteins that are identified during this analysis are called ‘false-positives’, since they are not part of the protein corona. Eliminating false positives is necessary for reliable characterization of the protein corona. | |
| Traditionally, researchers separated nanoparticle-protein complexes from unbound proteins using size exclusion chromatography, or repeated rounds of differential centrifugation and ‘washing’. Both of these techniques require that the nanomaterial-protein complexes be much larger and/or heavier than unbound proteins. In differential centrifugation, samples are spun at high speed, producing forces on the unbound proteins and protein-nanomaterial complexes that are often thousands of times the force of gravity. In response to these forces, large, heavy nanomaterial-protein complexes migrate to the bottom of a tube, while the lighter unbound proteins remain suspended. The unbound proteins can then be discarded, leaving only the nanoparticle-protein complexes. | |
| In the case of ‘ultrasmall’ nanomaterials (those with diameters of less than 10nm), the protein-nanomaterial complexes are about the same size and density as the proteins themselves. During differential centrifugation, the protein-nanomaterial complexes migrate at roughly the same rate as unbound proteins. In this case, it is not possible to isolate the nanoparticle-protein complexes. Without a suitable separation technique, researchers have been unable to study the composition of the protein corona formed around ‘ultrasmall’ nanomaterials. | |
| Reporting in the journal RSC Advances ("The protein corona of dendrimers: PAMAM binds and activates complement proteins in human plasma in a generation dependent manner"), a collaborative research team based at University College in Dublin, Ireland and Copenhagen University in Denmark have developed a new strategy to separate ultrasmall nanomaterial-protein complexes from unbound proteins. The team, led by Drs. Kenneth Dawson and Marité Cárdenas used electrophoresis to purify ultrasmall nanoparticle-protein complexes from blood plasma (the acellular, protein-rich component of blood). | |
| Dawson told Nanowerk in a recent interview that “the key is to get the proteins that are bound to separate from the ones that are not so that we are then able to characterize them. We wanted to introduce a different way of separating nanoparticle-protein complexes when it is not possible to use centrifugation or chromatographic techniques. We needed a new approach that would allow us to characterize the protein corona around ultrasmall nanoparticles such as dendrimers and quantum dots.” | |
| As opposed to differential centrifugation or size-exclusion chromatography, gel electrophoresis separates compounds based on differences in their ‘electrophoretic mobility’. Electrophoretic mobility is a measure of how fast a compound moves through a suspending medium when an electrical potential is applied to it, and depends on both the size and charge of the compound. Dawson, Cárdenas, and their team hypothesized that after exposing ultrasmall nanoparticles to blood plasma, the resulting nanoparticle-protein complexes would have an electrophoretic mobility that was different from unbound proteins. | |
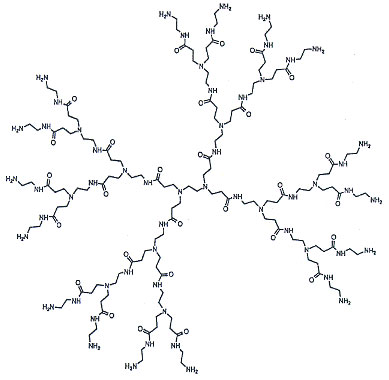
| Dawson and his colleagues used dendrimers as a model ultrasmall nanomaterial. Dendrimers are polymeric nanomaterials formed by sequentially connecting strands of a well-defined, branched polymer onto a central template (see Fig. 2). Dendrimers are classified by their ‘generation number’ which described how many sequential additions of the polymer unit have been made. In this study, Dawson and his colleagues used dendrimers built from poly(amido-amine) (PAMAM), with generation numbers ranging from 4 to 7 (corresponding to diameters ranging from 4 to 8nm). | |
 |
|
| Figure 2: Example of a generation 2 PAMAM dendrimer. (Image courtesy of Dendritech – www.dendritech.com) | |
| In addition to serving as a well-defined model system, dendrimers hold promise in a number of practical applications. PAMAM dendrimers, for example, are being developed as delivery vehicles for drugs or genes, or as components of larger nanoparticle assemblies. However, their propensity to activate the complement and coagulation pathways in blood has created a barrier to their clinical translation. Activation of these pathways can lead to anaphylaxis and/or thrombosis. Moreover, adsorption of complement and coagulation proteins can mediate the rapid removal of dendrimers from the blood stream by immune cells, which prevents them from accumulating at a target site. | |
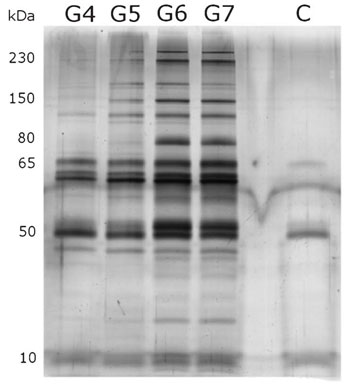
| The extent to which PAMAM dendrimers activate the complement and coagulation pathways increases as the generation number grows, suggesting that dendrimers of varying generation number interact with the protein components of these pathways in different ways. Dawson and his colleagues believe that these differences would translate into variations in the composition of the resulting protein corona. To test this theory, they incubated PAMAM dendrimers in blood plasma and separated the resulting dendrimer-protein complexes for unbound proteins using gel electrophoresis. They then used polyacrylamide gel electrophoresis and mass spectrometry to characterize the protein corona (see Fig. 3). | |
 |
|
| Figure 3: Image showing protein ‘bands’ associated with PAMAM dendrimers of varying generation number from polyacrylamide gel electrophoresis. Note how there are more bands present on the G6 and G7 lanes, and that these bands are darker than on the G4 and G5 lanes. (Reprinted with permission from Royal Society of Chemistry) | |
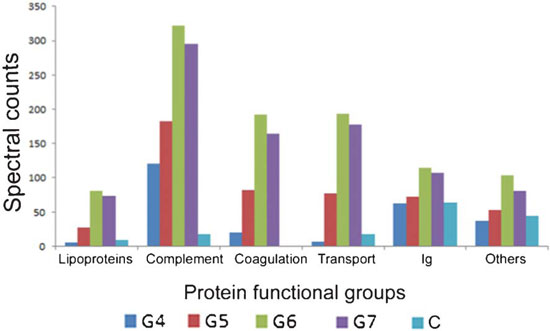
| Consistent with their theory, Dawson and his team found that lower-generation dendrimers adsorb fewer proteins and a different set of proteins as compared to their higher-generation counterparts. Higher-generation dendrimers adsorbed more proteins involved in the complement and coagulation cascades, which correlates with their greater tendency to activate these pathways in blood. Major components of the complement pathway, including complement factor C3 and C4, and key mediators of the coagulation pathway (fibrinogen & prothrombin) bound selectively to dendrimers with 6 or 7 generations (see Fig. 4). | |
 |
|
| Figure 4: Total spectral counts from proteins in each functional category adsorbed to dendrimers of varying generation number. Spectral counts give an indication as to how much protein is present in each functional category on the dendrimer surface. Note how the total amount of protein involved in complement activation and coagulation increase as dendrimer generation number increases, consistent with greater modulation of these pathways. (Reprinted with permission from Royal Society of Chemistry) | |
| “Activation of the complement and coagulation pathways is expected for cationic nanoparticles, like the dendrimers used here. What is interesting is that as you move below generation 4, you don’t see activation that is as strong. At the same time you see a shift in the proteins present within the corona.” explains Dawson. “This shows that ultrasmall nanoparticles interact with proteins, in particular those of the complement and coagulation pathways, in a way that is fundamentally different from analogous larger particles.” | |
| When a protein interacts with a nanoparticle that has a diameter that is much larger than the protein, the protein ‘sees’ the nanomaterial surface as essentially flat. However, as the nanoparticle diameter approaches the diameter of the protein, the local surface curvature of the nanoparticle becomes significant. This has the effect of altering the space on the nanomaterial that is available for the protein to interact with, and also altering the interaction of adjacent adsorbed proteins with each other. Both of these effects contribute to differences in the nature of the protein-nanomaterial interactions. This, in turn, leads to differences in the composition of the protein corona, as well as the extent to which the nanomaterials activate the complement and coagulation pathways. | |
| “There is a conceptual difference in the way the protein corona forms around ultrasmall nanomaterials. As the nanomaterial approaches the same size as the proteins, the proteins will begin to see the nanomaterial as if it were a protein itself” says Dawson. “The interactions become more specific. It is as if the nanoparticle becomes part of a larger protein complex.” | |
| In the case of the PAMAM dendrimers used in this study, it is not clear to what extent the observed differences in protein interactions are the result of differences in size and to what extent they are the result of differences in functional group density. As dendrimer generation number increases, the density of amine groups on the surface also increases. The increased functional group density may be, at least partially, the reason why higher-generation PAMAM dendrimers interact more strongly with blood proteins. To distinguish between these possibilities, the protein corona around other model ‘ultrasmall’ nanoparticles will need to be studied. | |
| It is likely that the gel electrophoresis strategy developed by Dawson and his colleagues can be adapted to study the protein corona formed around other types of ultrasmall nanoparticles (such as quantum dots). However, Dawson warns that the technique will need to be evaluated on a case-by-case basis. The differences in electrophoretic mobility between protein-nanomaterial complexes and free proteins may not hold for all nanomaterial types. | |
| As this study highlights, much of the increase in our understanding of the protein corona has come from technical advances that have enabled better purification and/or characterization of protein-nanomaterial complexes. Continued development in our understanding of the protein corona, how it depends on nanomaterial properties, and how it contributes to the ultimate biological response will be driven in large part by the development of more sophisticated characterization techniques. | |
| Moving forward, Dawson believes that it is not enough to look only at which proteins are present in the corona, but to also characterize their orientation and arrangement on the surface. Once this is established, it will be possible to look at how these properties influence the biological response including the activation and regulation of enzymatic pathways, cellular response, and toxicity. Developing these relationships will allow the biological response to a nanomaterial to be predicted by studying the protein corona, which will facilitate the design of nanomaterial formulations that are safer and more effective. | |
| By Carl Walkey, Integrated Nanotechnology & Biomedical Sciences Laboratory, University of Toronto, Canada. | |
|
Become a Spotlight guest author! Join our large and growing group of guest contributors. Have you just published a scientific paper or have other exciting developments to share with the nanotechnology community? Here is how to publish on nanowerk.com. |
|
