Nanopore DNA Sequencing: A Powerful Tool for Genetic Analysis
What is Nanopore DNA Sequencing?
Nanopore DNA sequencing is a cutting-edge technology that enables the direct, real-time analysis of long DNA or RNA fragments. Unlike conventional sequencing methods that require the amplification and fragmentation of DNA, nanopore sequencing allows for the sequencing of single molecules of DNA or RNA as they pass through a nanoscale pore. This innovative approach has revolutionized the field of genomics, offering high-throughput, low-cost, and portable sequencing solutions.
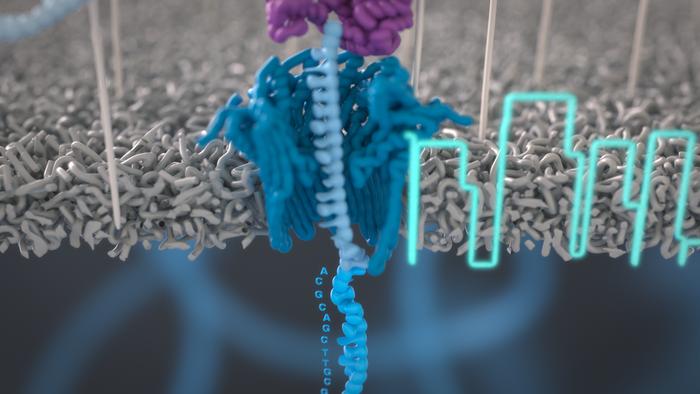
Key Components of Nanopore Sequencing
Nanopore sequencing relies on two main components: a nanopore and a detection system.
Nanopores
Nanopores are tiny holes with diameters in the range of 1-10 nanometers. They can be biological, such as protein pores like α-hemolysin or MspA, or solid-state, such as those made from silicon nitride or graphene. Biological nanopores offer high sensitivity and specificity, while solid-state nanopores provide stability and the ability to tune pore size and shape.
Detection System
The detection system in nanopore sequencing typically involves measuring the changes in electrical current as DNA or RNA passes through the nanopore. Each nucleotide (A, T, C, G) causes a characteristic disruption in the current, allowing for the identification of the DNA sequence. The electrical signal is recorded and processed in real-time, enabling the rapid determination of the nucleotide sequence.
Advantages of Nanopore Sequencing
Nanopore sequencing offers several advantages over traditional sequencing methods:
- Long Read Lengths: Nanopore sequencing can generate reads up to several hundred kilobases in length, enabling the analysis of long, complex genomic regions and the resolution of structural variations.
- Real-Time Sequencing: Nanopore sequencing allows for the real-time analysis of DNA or RNA, providing immediate access to sequencing data and enabling in-field or point-of-care applications.
- Minimal Sample Preparation: Nanopore sequencing requires minimal sample preparation, as it can directly sequence native DNA or RNA without the need for amplification or fragmentation.
- Portability: Nanopore sequencing devices are compact and portable, allowing for sequencing in remote or resource-limited settings.
Applications of Nanopore Sequencing
Nanopore sequencing has a wide range of applications in genomics and related fields:
Whole Genome Sequencing
Nanopore sequencing enables the rapid and cost-effective sequencing of entire genomes, including those of humans, animals, plants, and microorganisms. The long read lengths generated by nanopore sequencing facilitate the assembly of complex genomes and the identification of structural variations.
Transcriptome Analysis
Nanopore sequencing can be used for the direct sequencing of RNA, allowing for the analysis of alternative splicing, isoforms, and gene expression. This enables the study of the dynamic nature of the transcriptome and the identification of novel transcripts.
Epigenetic Analysis
Nanopore sequencing can detect DNA modifications, such as methylation, directly during sequencing. This capability enables the study of epigenetic regulation and its role in gene expression and disease.
Clinical Diagnostics
Nanopore sequencing has the potential to revolutionize clinical diagnostics by providing rapid, on-site sequencing for the identification of pathogens, the detection of genetic disorders, and the monitoring of disease progression. The portability and real-time nature of nanopore sequencing make it well-suited for point-of-care applications.
Limitations and Strategies for Improvement
While nanopore sequencing has revolutionized the field of genomics, it still faces several limitations, particularly in terms of accuracy. The accuracy of nanopore sequencing depends on various factors, such as the type of nanopore used, the quality of the sample, and the complexity of the genome being sequenced.
Biological Nanopores
Biological nanopores, such as α-hemolysin and MspA, offer high sensitivity and specificity due to their well-defined structure and the ability to engineer them for specific applications. However, they also have some limitations:
- Homopolymer Errors: Biological nanopores often struggle to accurately detect homopolymeric regions (stretches of identical nucleotides) due to the rapid passage of multiple identical bases through the pore.
- Limited Pore Stability: Biological nanopores are prone to degradation over time, which can affect the consistency and reliability of sequencing results.
- Influence of Experimental Conditions: The performance of biological nanopores can be influenced by factors such as pH, temperature, and salt concentration, requiring careful optimization of experimental conditions.
To address these limitations, researchers are exploring strategies such as the use of molecular adapters or modified bases to slow down the translocation of DNA through the pore, allowing for more accurate detection of homopolymers. Additionally, protein engineering approaches are being employed to improve the stability and robustness of biological nanopores.
Solid-State Nanopores
Solid-state nanopores, made from materials such as silicon nitride or graphene, offer several advantages over biological nanopores, including increased stability, tunable pore size and shape, and the potential for integration with electronic devices. However, they also have some limitations:
- Lower Sensitivity: Solid-state nanopores typically have lower sensitivity compared to biological nanopores, making it more challenging to detect subtle differences between nucleotides.
- Nonspecific Interactions: The surface of solid-state nanopores can interact nonspecifically with DNA, leading to signal noise and reduced accuracy.
- Fabrication Challenges: The precise fabrication of solid-state nanopores with consistent size and shape remains a challenge, affecting the reproducibility of sequencing results.
To overcome these limitations, researchers are investigating novel nanopore materials, such as two-dimensional nanomaterials (e.g., graphene, MoS2) and hybrid nanopores that combine the advantages of biological and solid-state nanopores. Surface modification strategies, such as the functionalization of nanopore surfaces with specific molecules, are being explored to reduce nonspecific interactions and improve signal-to-noise ratios.
Base-Calling Algorithms and Error Correction
In addition to nanopore development, advances in base-calling algorithms and error correction methods are crucial for improving the accuracy of nanopore sequencing. Machine learning techniques, such as deep learning neural networks, are being applied to develop more sophisticated base-calling algorithms that can accurately interpret the complex electrical signals generated during sequencing.
Error correction strategies, such as consensus sequencing and hybrid sequencing approaches that combine nanopore sequencing with short-read sequencing technologies, are also being employed to improve the overall accuracy of sequencing results. By leveraging the strengths of multiple sequencing technologies, hybrid approaches can provide a more comprehensive and accurate view of the genome.
As research continues to address these limitations and develop innovative solutions, the accuracy and reliability of nanopore sequencing are expected to improve, further expanding its applications in various fields, from basic research to clinical diagnostics.
Further Reading
Nature Biotechnology, Nanopore sequencing technology, bioinformatics and applications
