| Apr 05, 2022 |
Engineering single-molecule DNA computing with magnetic tweezers
(Nanowerk News) Deoxyribonucleaic acid, DNA, can store vast amounts of information encoded as sequences of the molecules, known as nucleotides, cytosine (C), guanine (G), adenine (A), or thymine (T). The complexity and enormous variance of different species’ genetic codes demonstrates how much information can be stored within DNA encoded using CGAT, and this capacity can be put to use in computing.
|
|
Specifically, DNA molecules can be used to process information, using a bonding process between DNA pairs known as hybridization. This takes single strands of DNA as input and produces subsequent strands of DNA through transformation as output.
|
|
That means that rather than electrons flowing in and out of electronic transistors, DNA-based logic gates receive and produce molecules as signals.
|
|
DNA computing was first demonstrated in 1994 by Leonard Adleman who encoded and solved the traveling salesman problem, a maths problem to find the most efficient route for a salesman to take between hypothetical cities, entirely in DNA.
|
|
Over the past two decades, various DNA nanotechnology methods have been developed to perform Boolean logic computing using DNAzyme or enzyme cleavage, light irradiation, and DNA self-assembly.
|
|
However, most of these methods have been conducted at the bulk level, which may lead to missing information and be less controllable.
|
|
In new work, reported in Nano Letters ("Single-Molecule Resettable DNA Computing via Magnetic Tweezers"), scientists engineered single-molecule DNA computing controlled by stretching forces using magnetic tweezers. By tracking the real-time signals of the DNA extension, the output can be determined at a single base-pair resolution.
|
|
This real-time tracking, label-free, and force-controlled single-molecule DNA computing shows potential in precisely modulating the computing processes, which could give new insight into the design of molecular computing.
|
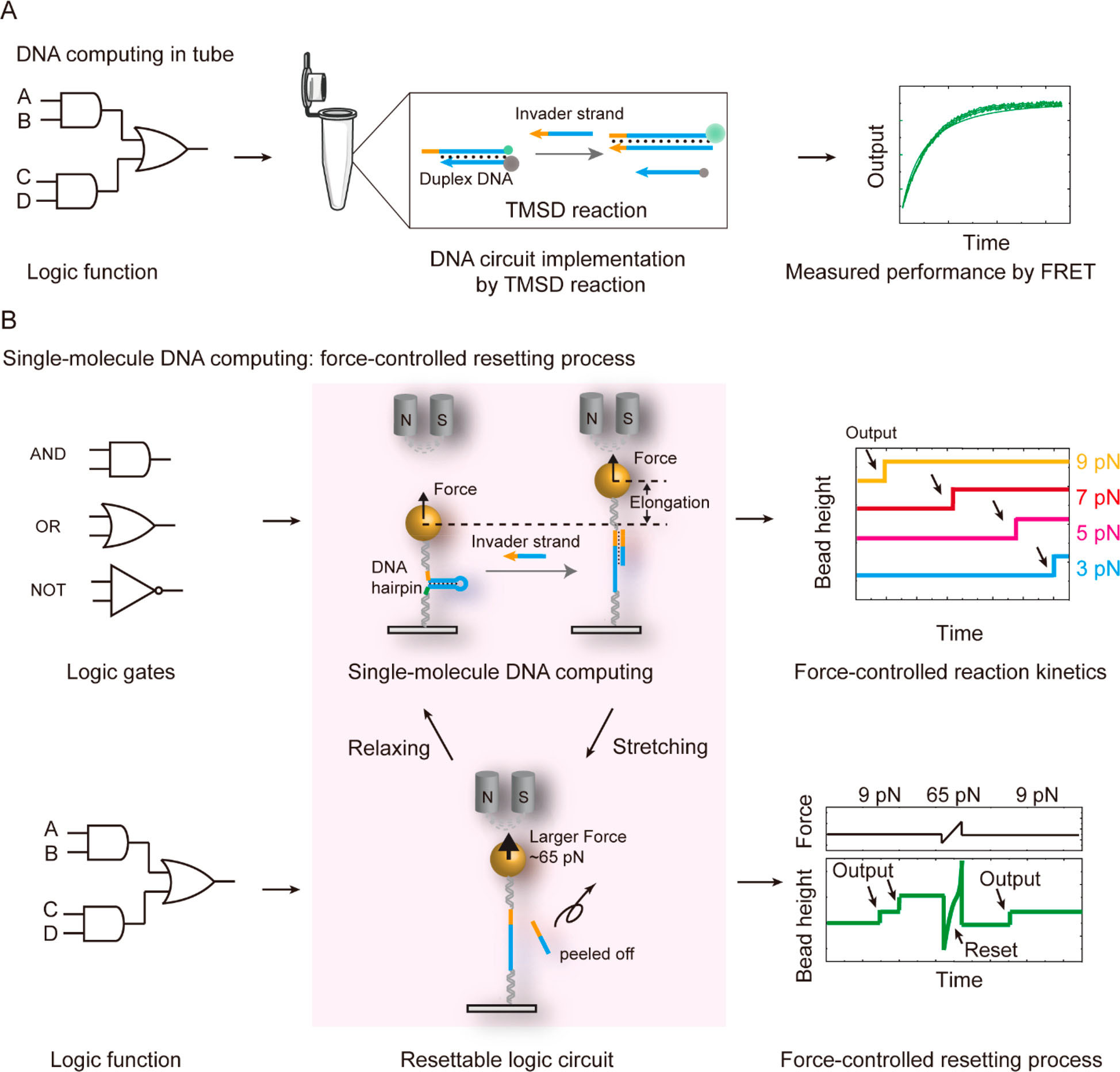 |
| Schematic illustration of a force-controlled resettable single-molecule DNA computing system. (A) Traditional DNA computing measurement in a tube. Generally, the output signal is determined by optical spectroscopy. (B) New method for realizing DNA computing by force spectroscopy using magnetic tweezers. The processes of single-molecule DNA logic gates and circuits can be controlled and restarted by stretching forces. The step like jump in the time trace of bead height represents the output signal. (Reprinted with permission by American Chemical Society) (click on image to enlarge)
|
|
As the team reports, due to the real-time tracking ability, a toehold-mediated DNA strand displacement (TMSD) event can be determined by a step like hairpin unfolding signal with a base-pair resolution. Depending on the stretching forces, the reaction rate of the basic TMSD reaction can be accelerated exponentially.
|
|
They also used single-molecule OR, AND, and NOT gates under the stretching force. Although adding some of the DNA strands in the logic gates may not generate the output signal in a single trace, repeating the stretching cycles can give accurate probabilities.
|
|
Moreover, they proved that the TMSD reaction in logic gates and circuits can be stopped and reset by repeating the stretching cycles.
|

 By
Michael
Berger
– Michael is author of three books by the Royal Society of Chemistry:
Nano-Society: Pushing the Boundaries of Technology,
Nanotechnology: The Future is Tiny, and
Nanoengineering: The Skills and Tools Making Technology Invisible
Copyright ©
Nanowerk LLC
By
Michael
Berger
– Michael is author of three books by the Royal Society of Chemistry:
Nano-Society: Pushing the Boundaries of Technology,
Nanotechnology: The Future is Tiny, and
Nanoengineering: The Skills and Tools Making Technology Invisible
Copyright ©
Nanowerk LLC
