| Posted: September 26, 2008 |
Structural 'snapshots' of a protein implicated in Alzheimer's disease |
|
(Nanowerk News) A recent study describes the structure of the active form of BACE1, which is an enzyme implicated in Alzheimer’s disease. BACE1 cleaves amyloid precursor protein (APP), thereby releasing amyloid β peptide (Aβ), the primary component of amyloid plaques found in the brains of patients with Alzheimer’s disease.
|
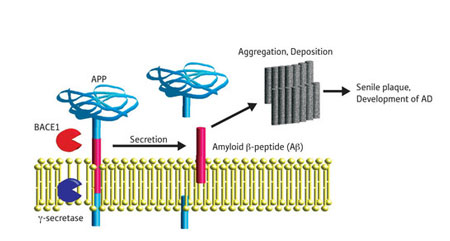 |
| Schematic depicting generation of Aβ from APP. γ-secretase is another enzyme involved in the process. AD, Alzheimer’s disease. (Image: RIKEN)
|
|
As amyloid plaques are thought by many to inflict brain cell damage that results in Alzheimer’s disease, efforts are under way to design drugs to inhibit the activity of BACE1. Complicating these efforts is the fact that BACE1 seems to cleave APP in vesicles called endosomes, which sport a pH much more acidic than that of other areas of the cell or the extracellular fluid.
|
|
Structures of several BACE1 complexes have been solved using a technique called x-ray crystallography, wherein structural information is gleaned from x-rays diffracted from crystallized versions of proteins. However, never before has a structural view of active BACE1 been available. In a paper recently published in Molecular and Cellular Biology, Nobuyuki Nukina and colleagues from the RIKEN Brain Science Institute in Wako and the RIKEN SPring-8 Center in Harima present and analyze crystals of active BACE1 ("Crystal Structure of an Active Form of BACE1, an Enzyme Responsible for Amyloid β Protein Production").
|
|
To identify conditions in which crystallized BACE1 is active, the researchers soaked BACE1 crystals in acidic (pH 4.0, 4.5 and 5.0) and neutral (pH 7.0) solutions, together with synthetic APP peptides engineered to fluoresce after cleavage. In agreement with data localizing BACE1 activity to acidic endosomes, crystallized BACE1 cleaved APP at acidic but not neutral pH.
|
|
Comparative analyses revealed substantial differences in the shape of BACE1 crystals soaked in acidic and neutral solutions, suggesting that BACE1 undergoes structural rearrangements during activation. Most notable was the position of the ‘flap’ covering the active site of BACE1, which was open and closed in acidic and neutral crystals, respectively. Also observed were marked changes in the shape of the BACE1 site at which the substrate—in this case, APP—binds.
|
|
Binding of a water molecule—thought to be important in the chemical reaction through which BACE1 cleaves APP—became weaker as the pH was lowered. Whether BACE1 exists as a mix of hydrated active and dehydrated inactive forms in endosomes remains unclear.
|
|
These findings highlight the importance of considering environmental factors such as pH in structure-based design of enzyme inhibitors. “The structure of the active form of BACE1 identified here should be used for developing drugs to regulate Aβ production,” says Nukina.
|

