| Posted: Jul 17, 2014 | |
DNA sequencing reaches new lengths |
|
| (Nanowerk Spotlight) Sequencing technologies have made it cheaper and faster to read the sequence of bases on a strand of DNA. A promising technology to take these advances further is nanopore sequencing. Individual strands of DNA are moved through a nanopore gap not much wider than the DNA itself. As the DNA passes through the nanopore, continuous information is gained about the sequence of individual bases – the A, C, G and Ts that make up DNA. So far the technology has been used for sequencing relatively short fragments of DNA around 100 bases long. | |
| “One reason why people are so excited about nanopore DNA sequencing is that the technology could possibly be used to create ‘tricorder’-like devices for detecting pathogens or diagnosing genetic disorders rapidly and on-the-spot,” commented Andrew Laszlo, a researcher in nanopore technology at the University of Washington. | |
| Researchers from the University of Washington’s Departments of Physics and Genome Sciences have developed a nanopore sequencing technique reaching read lengths of several thousand bases. The result is the latest in a series of advances in nanopore technology developed at the university. | |
| The team, led by Jens Gundlach, published their findings in Nature Biotechnology as an advanced online publication on June 25, 2014 ("Decoding long nanopore sequencing reads of natural DNA"). | |
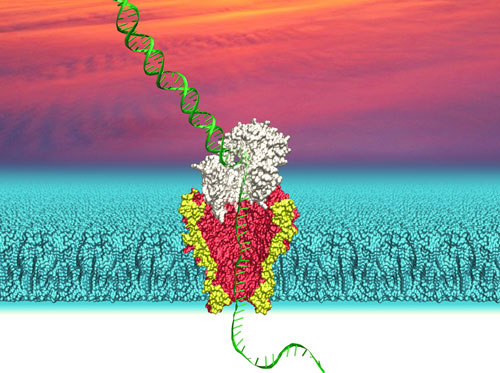 |
|
| Depiction of DNA (green) passing through a polymerase (white) followed by the nanopore protein MspA (yellow/red) embedded in a lipid bilayer (cyan). University of Washington researchers use this system to identify the sequences of individual strands of DNA. (Image: Ian Derrington, University of Washington) | |
| “This is the first time anyone has shown that nanopores can be used to generate interpretable signatures corresponding to very long DNA sequences from real-world genomes,” said co-author Jay Shendure, an associate professor in Genome Sciences, “It’s a major step forward.” | |
| The idea for nanopore sequencing originated in the 90s: a lipid membrane, similar to the material that makes up the cell wall, acts as a barrier separating two liquids. Inserted into the membrane is a tiny gap, just nanometers across, called a nanopore. By applying a voltage difference across the barrier, ions in the liquid try to move between the two sides of the barrier and the only way to do this is to flow through the nanopore. The movement of the charged molecules between the two liquids is a current, just like electrons moving along a wire in an electrical circuit, and can be recorded. | |
| Any DNA in the system is also pulled towards the other side of the barrier by the voltage difference, since DNA is negatively charged, and just like the ions it has to pass through the nanopore. The difference is that the DNA is much bigger than the ions and partially blocks the nanopore, making it harder for the smaller molecules to pass through. As the ions are blocked by the DNA, there is a measurable difference in the current flowing across the membrane which is dependent on the DNA base passing through the nanopore. By measuring the changing current, information can be gained on the bases passing through. | |
| The researchers created the nanopore by inserting a single protein called Mycobacterium smegmatis porin A, or MspA, in the membrane. MspA is normally found lining the membrane of a species of bacteria, controlling the intake of nutrients. | |
| One challenge the researchers faced was the control of the DNA passing through the nanopore. Normally, the DNA would zip through the MspA nanopore too fast to detect the changes in the current. The researchers slowed the DNA movement through the pore using a second protein called phi29 DNA polymerase (DNAP), which captures DNA and slows its movement through the pore. | |
| The shape of the protein MspA meant that several bases passed through the nanopore at one time and the current changes were the result of a combination of those bases. This presented another challenge. Since several bases passed through the nanopore at one time, the researchers needed a way to decipher what the current changes meant. To do this, they first made a library of DNA sequences that contains all possible combinations of 4 nucleotides (for the mathematically inclined, the library is 44 = 256 bases long – a string of 4 bases with 4 possible choices for each DNA base). The library, whose sequence was already known, was run though the nanopore first to find the current associated with each set of DNA base combinations. They combined the library measurements with known genome sequences to generate a set of expected current changes that could be compared to experimental measurements. | |
| The researchers tested their approach by sequencing the entire genome of bacteriophage Phi X 174, a virus that infects bacteria and is used as a benchmark for evaluating new sequencing technologies. The impressive feat here is the length of the genome they sequenced – the Phi X 174 genome is 4,500 bases long. Other nanopore technologies have been limited to sequencing DNA fragments that were much shorter. | |
| “Despite the remaining hurdles, our demonstration that a low-cost device can reliably read the sequences of naturally occurring DNA and can interpret DNA segments as long as 4,500 nucleotides in length represents a major advance in nanopore DNA sequencing,” explained Gundlach. | |
|
By Dr Richard Muscat (on Twitter: @RAMuscat), Molecular Engineering and Sciences Institute, University of Washington
|
|
|
Become a Spotlight guest author! Join our large and growing group of guest contributors. Have you just published a scientific paper or have other exciting developments to share with the nanotechnology community? Here is how to publish on nanowerk.com. |
|
