| Posted: Oct 05, 2009 | |
Nanotechnology sensor detects living bacteria at ultralow concentrations |
|
| (Nanowerk Spotlight) A pathogen is a an organism (bacterium, virus, parasite) that causes disease in another organism. According to data from the U.S. Centers for Disease Control and Prevention (CDC), known pathogens account for an estimated 14 million illnesses, 60,000 hospitalizations, and 1,800 deaths each year in the United States. In addition to the food supply, microbacterial infections are a major issue in hospitals. Other CDC data estimates that infections acquired in hospitals alone affect approximately 2 million persons annually. In the U.S., between 44,000 and 98,000 people die every year from infections they picked up in hospitals. The numbers for Europe are equally scary. | |
| Add to that the potential for terrorist attacks with pathogen-based bioweapons and it becomes clear that the development of fast and reliable biosensors is a major issue. The problem today is that analytical tools for the rapid detection of pathogens tend to be slow. | |
| Traditionally, identifying a pathogen such as E. coli, Salmonella or Listeria requires cell culturing, which takes time – time that often means more contamination and illnesses or even deaths. Here is an example from the FDA's recommended method for determining E. coli: | |
| Weigh 50 g food into sterile high-speed blender jar. Add 450 mL of Butterfield's phosphate-buffered water and blend for 2 min. Prepare decimal dilutions with sterile Butterfield's phosphate diluent. Number of dilutions to be prepared depends on anticipated coliform density. Shake all suspensions 25 times in 30 cm arc or vortex mix for 7 s. Do not use pipets to deliver <10% of their total volume. Transfer 1 mL portions to 3 LST tubes for each dilution for at least 3 consecutive dilutions. Hold pipet at angle so that its lower edge rests against the tube. Let pipet drain 2-3 s. Not more than 15 min should elapse from time the sample is blended until all dilutions are inoculated in appropriate media. Incubate LST tubes at 35°C. Examine tubes and record reactions at 24 ± 2 h for gas, i.e., displacement of medium in fermentation vial or effervescence when tubes are gently agitated. Re-incubate gas-negative tubes for an additional 24 h and examine and record reactions again at 48 ± 2 h. Perform confirmed test on all presumptive positive tubes (which takes another 2 days). | |
| Researchers have managed to develop much faster detection systems based on nanotechnology (see for instance: Nanotechnology barcodes to quickly identify biological weapons). The fastest assays however, those that have reaction times in the minutes range, require pretreatment steps to condition the test samples and to perform cell lysis to extract the suitable target DNA. A major drawback which complicates testing considerably. | |
| In their effort to develop a fast, sensitive, selective, inexpensive, and easy-to-use method for detecting and quantifying pathogenic bacterial cells, researchers in Spain have now demonstrated a carbon nanotube based potentiometric biosensor for selectively detecting one single colony-forming unit of the bacterium Salmonella Typhi in close to real time. | |
 |
|
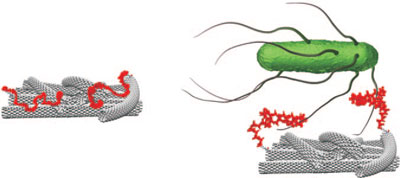
| An aptamer attached to an electrode coated with single-walled carbon nanotubes interacts selectively with bacteria. The resulting electrochemical response is highly accurate and reproducible and starts at ultralow bacteria concentrations, providing a simple, selective method for pathogen detection. (Reprinted with permission from Wiley-VCH Verlag) | |
| According to F. Xavier Rius, a professor at the Rovira i Virgili University in Tarragona, Spain, who heads the Chemometrics, Qualimetrics and Nanosensors Group, "the most important strength of this biosensor is that simple positive/negative tests can be carried out in real zero-tolerance conditions and without cross reaction with other types of bacteria. The ease with which measurements are taken in potentiometric analysis opens the door to greater simplicity in microbiological analysis." | |
| To build their sensor, the Spanish team linked carboxylated single-walled carbon nanotubes (SWCNT) to an aptamer. Aptamers are not only highly suitable receptors for the selective and high-proficiency detection of a wide range of molecular targets, including bacteria, they have also shown to self-assemble on carbon nanotubes. This hybrid nanomaterials has already been demonstrated as effective nanobiosensors ("Detection and Titer Estimation of Escherichia coli Using Aptamer-Functionalized Single-Walled Carbon-Nanotube Field-Effect Transistors"). | |
| The hybrid material aptamer-SWCNT acts as both the sensing and the transducing layer of the biosensor. The presence of target bacteria promotes a conformational change in the aptamer that separates the phosphate groups, largely ionized at pH 7.4, from the SWCNT sidewalls, inducing a charge change to the SWCNT and the subsequent change of the recorded potential. Ruiz' team also found that their biosensors show a high degree of electivity, i.e. no response was shown for bacteria other than the target bacterium Salmonella Typhi. | |
| It appears that this new biosensor makes the detection of pathogens as easy as measuring the pH value. | |
| Ruiz and team have reported their findings in a recent issue of Angewandte Chemie International Edition ("Immediate Detection of Living Bacteria at Ultralow Concentrations Using a Carbon Nanotube Based Potentiometric Aptasensor"). | |
 By
Michael
Berger
– Michael is author of three books by the Royal Society of Chemistry:
Nano-Society: Pushing the Boundaries of Technology,
Nanotechnology: The Future is Tiny, and
Nanoengineering: The Skills and Tools Making Technology Invisible
Copyright ©
Nanowerk LLC
By
Michael
Berger
– Michael is author of three books by the Royal Society of Chemistry:
Nano-Society: Pushing the Boundaries of Technology,
Nanotechnology: The Future is Tiny, and
Nanoengineering: The Skills and Tools Making Technology Invisible
Copyright ©
Nanowerk LLC
|
|
|
Become a Spotlight guest author! Join our large and growing group of guest contributors. Have you just published a scientific paper or have other exciting developments to share with the nanotechnology community? Here is how to publish on nanowerk.com. |
|
