| Posted: Aug 21, 2017 | |
Amplifying fluorescent signals in molecular sensing with DNA walkers |
|
| (Nanowerk Spotlight) Synthetic nanomotors and DNA walkers, which mimic a cell's transportation system, are intricately designed systems that draw chemical energy from the environment and convert it into mechanical motion. | |
| This kind of engineered molecular motors are a prime example of nanotechnology's efforts to imitate biology in order to build artificial chemical systems with similar power and capabilities as living cells. Natural molecular motors are also studied as fundamental building blocks of bottom-up nanotechnology, for instance in nanorobots or in programmable molecular assembly lines. | |
| "Using DNA walkers as signal amplifier for nucleic acids detection has only recently been reported," Prof. Dr. Philip Tinnefeld from the Institute of Physical and Theoretical Chemistry at Technical University (TU) Braunschweig, tells Nanowerk. "Inspired by these findings, we converted a nicking enzyme-assisted DNA walker into a linear fluorescence signal amplifier on a rectangle DNA origami that can improve the detection of target molecules such as nucleic acids." | |
| As Tinnefeld and his team reported in Nano Letters ("A DNA Walker as a Fluorescence Signal Amplifier"), the DNA walker counts the number of walking steps which is typically around 20-60 steps. | |
 |
|
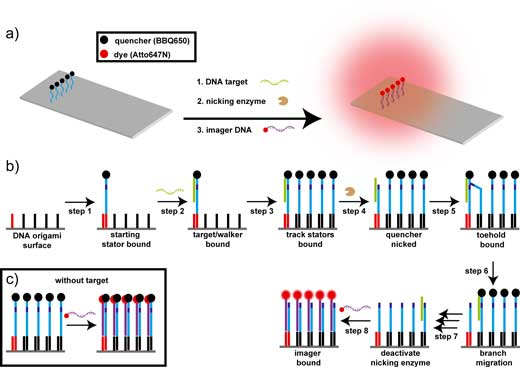
| (a) Scheme of the DNA walker on the DNA origami. The track for the DNA walker is formed by extended staple strands in the DNA origami and hybridizing quencher labeled stator strands on top of the DNA origami. (b) Fluorescence amplification mechanism of the DNA walker. The start site is marked by first hybridizing a single start stator (step 1) followed by target binding (step 2). The preparation of the DNA origami is finalized by hybridizing the remaining stators with quenchers (step 3; here 4 additional stator strands). The fluorescence amplification experiment is launched with the addition of the nicking enzyme (step 4) that cleaves the stator strand at a position marked by the target/stator duplex. After nicking, the walker proceeds to the next stator via toehold mediated branch migration (step 5 and 6). The autonomous walking steps are repeated until the end of the track is reached (step 7). Inactivating the enzyme and adding the imager strand finalizes the experiment (step 8). The brightness of the unquenched fluorophores corresponds to the number of steps the walker has moved. (c) In the absence of target, the imager strands can also bind to the stators, but their fluorescence is quenched, as the quenchers were not removed in the walking process. (Reprinted with permission by American Chemical Society) (click on image to enlarge) | |
| "From a biosensing point of view, we converted a target that in common assays creates a single-dye signal by being a binding partner into a catalyst that creates many signaling molecules," he notes. "Another unique feature is that this catalysis proceeds locally on a single DNA origami and the course of the reaction is therefore less dependent on diffusing molecules in solution." | |
| The walker can be combined with plasmonic effects for potential synergetic signal enhancement and be applied in molecular diagnostics as signal amplification mechanism. | |
| The researchers also find the walker is highly dependent on the correct sequence of target DNA and the rate constant for walking decreases by almost a factor of 2 for a single mismatch. Therefore the DNA walker system is suitable for sensitive DNA detection and single nucleotide polymorphisms (SNP) detection, which is important for clinical diagnostics. | |
| "While other signal enhancement mechanisms such as the polymerase chain reaction (PCR) can produce exponential signal growth compared to our linear fluorescence amplification, there is an interesting notion about the linear signal amplification," Tinnefeld points out. "The walker only amplifies linearly but therefore the correct sequence is examined for every step. This might facilitate detection mechanisms circumventing the need for PCR." | |
| Currently, the scientists at TU Braunschweig are trying to exploit different aspects of DNA nanotechnology for biosensing. As part of this work, they explore different signal amplification mechanisms such as plasmonic fluorescence enhancement and signal amplification by DNA walking. | |
| Going forward, they will try to build on their findings to detect low amounts of nucleic acids on consumer-type devices such as a modified smartphone. | |
| Tinnefeld's research group already has received funding for the interdisciplinary project POCEMON (point-of-care diagnostics with single molecule detection), in which they are going to develop a smartphone-based diagnostic point-of-care device. | |
| "The fact that each DNA origami walker creates a signal significantly higher than the background makes the approach appealing for sensitive DNA detection with single-nucleotide discrimination," he concludes. "This is taking DNA walkers on DNA nanostructures one step further from academic models of molecular motility to useful nanodevices." | |
 By
Michael
Berger
– Michael is author of three books by the Royal Society of Chemistry:
Nano-Society: Pushing the Boundaries of Technology,
Nanotechnology: The Future is Tiny, and
Nanoengineering: The Skills and Tools Making Technology Invisible
Copyright ©
Nanowerk LLC
By
Michael
Berger
– Michael is author of three books by the Royal Society of Chemistry:
Nano-Society: Pushing the Boundaries of Technology,
Nanotechnology: The Future is Tiny, and
Nanoengineering: The Skills and Tools Making Technology Invisible
Copyright ©
Nanowerk LLC
|
|
|
Become a Spotlight guest author! Join our large and growing group of guest contributors. Have you just published a scientific paper or have other exciting developments to share with the nanotechnology community? Here is how to publish on nanowerk.com. |
|
