Showing Spotlights 41 - 48 of 68 in category All (newest first):
 A very promising field of nanomotor research are DNA nanomachines. These are synthetic DNA assemblies that switch between defined molecular shapes upon stimulation by external triggers. They can be controlled by a variety of methods that include pH changes and the addition of other molecular components, such as small molecule effectors, proteins and DNA strands. Researchers have now designed and built a simple DNA machine that is capable of continuous rotation with controlled speed and direction - a function that might be very useful for example for molecular transport. This machine is driven by an externally controlled electric field. When this field is oscillated between four directions, it continuously reorients a rotor DNA that is asymmetrically attached to a DNA axle.
A very promising field of nanomotor research are DNA nanomachines. These are synthetic DNA assemblies that switch between defined molecular shapes upon stimulation by external triggers. They can be controlled by a variety of methods that include pH changes and the addition of other molecular components, such as small molecule effectors, proteins and DNA strands. Researchers have now designed and built a simple DNA machine that is capable of continuous rotation with controlled speed and direction - a function that might be very useful for example for molecular transport. This machine is driven by an externally controlled electric field. When this field is oscillated between four directions, it continuously reorients a rotor DNA that is asymmetrically attached to a DNA axle.
Jan 12th, 2010
 One challenge in designing nanomachines is being able to establish how well they work and optimize their performance. This is where single molecule techniques will play an important role. With advances in nanotechnologies, it is possible to construct simple nanomachines that can perform simple functions such as opening and closing of a DNA device (e.g. DNA tweezers or DNA switches), small rotational and translational motors and energy transfer cascades. Using single-molecule techniques researchers can watch individual nanomachines working and determine the functionality of their design. Researchers in Germany now have incorporated optical addressability to these nanomachines. Hence, they can optically detect and eventually control the state of the nanodevice.
One challenge in designing nanomachines is being able to establish how well they work and optimize their performance. This is where single molecule techniques will play an important role. With advances in nanotechnologies, it is possible to construct simple nanomachines that can perform simple functions such as opening and closing of a DNA device (e.g. DNA tweezers or DNA switches), small rotational and translational motors and energy transfer cascades. Using single-molecule techniques researchers can watch individual nanomachines working and determine the functionality of their design. Researchers in Germany now have incorporated optical addressability to these nanomachines. Hence, they can optically detect and eventually control the state of the nanodevice.
Dec 23rd, 2009
 Much of today's genetic research and diagnostics uses tools and technologies enabled by DNA's ability to bind to its complementary strand in a sequence specific manner. For biologists studying molecular mechanisms inside cells, this information helps to quantify the expression levels of genes. Detection of the binding - or hybridization - of DNA strands is at the heart of modern medicine. The technology for detecting DNA hybridization mainly relies on the use of fluorescent labels. The complementary strand coming from the sample bears a label, so detection of florescence signal indicates hybridization. While this may sound straightforward, it has major limitations. Researchers have now reported a new technique for genetic analysis using nanomechanical response of hybridized DNA/RNA molecules. This new technique is several orders of magnitude more sensitive than other approaches and it is a lot simpler to use.
Much of today's genetic research and diagnostics uses tools and technologies enabled by DNA's ability to bind to its complementary strand in a sequence specific manner. For biologists studying molecular mechanisms inside cells, this information helps to quantify the expression levels of genes. Detection of the binding - or hybridization - of DNA strands is at the heart of modern medicine. The technology for detecting DNA hybridization mainly relies on the use of fluorescent labels. The complementary strand coming from the sample bears a label, so detection of florescence signal indicates hybridization. While this may sound straightforward, it has major limitations. Researchers have now reported a new technique for genetic analysis using nanomechanical response of hybridized DNA/RNA molecules. This new technique is several orders of magnitude more sensitive than other approaches and it is a lot simpler to use.
Dec 21st, 2009
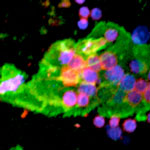 Bionanotechnology researchers are experimenting with techniques for attaching DNA nanoarrays to cell surfaces for various reasons: to label cell surfaces with functionalized micrometer-sized patches; to deliver materials such as nanoparticles or carbon nanotubes to cell surfaces; to deliver nucleic acids into the cell for gene silencing; or to engineer microtissues of cell/cell networks by using self-hybridizing properties of single stranded DNA molecules. A team of scientists in California has now successfully attached self-assembled DNA structures to cancer cells using two different methods. This is one of the first illustrations of the biomedical relevance of DNA arrays.
Bionanotechnology researchers are experimenting with techniques for attaching DNA nanoarrays to cell surfaces for various reasons: to label cell surfaces with functionalized micrometer-sized patches; to deliver materials such as nanoparticles or carbon nanotubes to cell surfaces; to deliver nucleic acids into the cell for gene silencing; or to engineer microtissues of cell/cell networks by using self-hybridizing properties of single stranded DNA molecules. A team of scientists in California has now successfully attached self-assembled DNA structures to cancer cells using two different methods. This is one of the first illustrations of the biomedical relevance of DNA arrays.
Sep 22nd, 2009
 A Japanese-UK research team has now demonstrated that cryoEM image analysis may be exploited to obtain structural information of sufficient resolution to reveal the absolute three-dimensional (3D) configuration of a designed DNA nanostructure. With their technique they have obtained structural information at sufficient resolution to visualize the DNA helix and reveal the absolute stereochemistry of a self-assembled DNA tetrahedron. Each edge is a 7 nm, 20 base pair duplex, and the edges are connected covalently through single unpaired adenosine nucleotides, making it a rigid, triangulated structure that could serve as a building block for larger 3D structures or as a molecular cage. This DNA tetrahedron is the smallest 3D nanostructure made by DNA self-assembly.
A Japanese-UK research team has now demonstrated that cryoEM image analysis may be exploited to obtain structural information of sufficient resolution to reveal the absolute three-dimensional (3D) configuration of a designed DNA nanostructure. With their technique they have obtained structural information at sufficient resolution to visualize the DNA helix and reveal the absolute stereochemistry of a self-assembled DNA tetrahedron. Each edge is a 7 nm, 20 base pair duplex, and the edges are connected covalently through single unpaired adenosine nucleotides, making it a rigid, triangulated structure that could serve as a building block for larger 3D structures or as a molecular cage. This DNA tetrahedron is the smallest 3D nanostructure made by DNA self-assembly.
Aug 25th, 2009
 DNA origami, tiny shapes and patterns self-assembled from DNA, have been heralded as a potential breakthrough for the creation of nanoscale circuits and devices. One roadblock to their use has been that they are made in solution, and they stick randomly to surfaces - like a deck of playing cards thrown onto a floor. Random arrangements of DNA origami are not very useful; if they carry electronic circuits for example, they are difficult to find and 'wire-up' into larger circuits. A collaboration between Caltech and IBM research Almaden has found a way to position and orient DNA origami on surfaces by creating sticky patches in the shape of origami - as a demonstration they positioned and aligned triangular DNA origami on triangular sticky patches. This success knocks down one of the major roadblocks for the use of DNA origami in computer nanotechnology.
DNA origami, tiny shapes and patterns self-assembled from DNA, have been heralded as a potential breakthrough for the creation of nanoscale circuits and devices. One roadblock to their use has been that they are made in solution, and they stick randomly to surfaces - like a deck of playing cards thrown onto a floor. Random arrangements of DNA origami are not very useful; if they carry electronic circuits for example, they are difficult to find and 'wire-up' into larger circuits. A collaboration between Caltech and IBM research Almaden has found a way to position and orient DNA origami on surfaces by creating sticky patches in the shape of origami - as a demonstration they positioned and aligned triangular DNA origami on triangular sticky patches. This success knocks down one of the major roadblocks for the use of DNA origami in computer nanotechnology.
Aug 18th, 2009
 Various forms of hyperthermia - a form of cancer treatment with elevated temperature in the range of 41-45C - have been intensively developed for the past few decades to provide cancer clinics with more effective and advanced cancer therapy techniques. The recent use of nanomaterials has shown promising for developing more effective hyperthermia agents. While most nanomedical hyperthermia research is conducted with various nanoparticles, carbon nanotubes are also of interest in these thermal ablation applications. So far, however, the utility of carbon nanotubes for in vivo use has been limited by self-association - i.e. they stick to each other. A new study has now demonstrated that DNA-encasement of multi-walled carbon nanotubes (MWCNTs) results in well-dispersed, single MWCNTs that are soluble in water and that display enhanced heat production efficiency relative to non-DNA-encased MWCNTs.
Various forms of hyperthermia - a form of cancer treatment with elevated temperature in the range of 41-45C - have been intensively developed for the past few decades to provide cancer clinics with more effective and advanced cancer therapy techniques. The recent use of nanomaterials has shown promising for developing more effective hyperthermia agents. While most nanomedical hyperthermia research is conducted with various nanoparticles, carbon nanotubes are also of interest in these thermal ablation applications. So far, however, the utility of carbon nanotubes for in vivo use has been limited by self-association - i.e. they stick to each other. A new study has now demonstrated that DNA-encasement of multi-walled carbon nanotubes (MWCNTs) results in well-dispersed, single MWCNTs that are soluble in water and that display enhanced heat production efficiency relative to non-DNA-encased MWCNTs.
Aug 13th, 2009
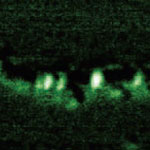 It its more than 25 years of existence, Scanning Tunneling Microscopy has predominantly brought us extremely detailed images of matter at the molecular and atomic level. The Scanning Tunneling Microscope (STM) is a non-optical microscope that scans an electrical probe over a surface to be imaged to detect a weak electric current flowing between the tip and the surface. It allows scientists to visualize regions of high electron density and hence infer the position of individual atoms and molecules on the surface of a lattice. Now, researchers in Japan have managed to partially sequence a single DNA molecule with a STM - a significant step towards the realization of electronic-based single-molecule DNA sequencing.
It its more than 25 years of existence, Scanning Tunneling Microscopy has predominantly brought us extremely detailed images of matter at the molecular and atomic level. The Scanning Tunneling Microscope (STM) is a non-optical microscope that scans an electrical probe over a surface to be imaged to detect a weak electric current flowing between the tip and the surface. It allows scientists to visualize regions of high electron density and hence infer the position of individual atoms and molecules on the surface of a lattice. Now, researchers in Japan have managed to partially sequence a single DNA molecule with a STM - a significant step towards the realization of electronic-based single-molecule DNA sequencing.
Aug 12th, 2009
 A very promising field of nanomotor research are DNA nanomachines. These are synthetic DNA assemblies that switch between defined molecular shapes upon stimulation by external triggers. They can be controlled by a variety of methods that include pH changes and the addition of other molecular components, such as small molecule effectors, proteins and DNA strands. Researchers have now designed and built a simple DNA machine that is capable of continuous rotation with controlled speed and direction - a function that might be very useful for example for molecular transport. This machine is driven by an externally controlled electric field. When this field is oscillated between four directions, it continuously reorients a rotor DNA that is asymmetrically attached to a DNA axle.
A very promising field of nanomotor research are DNA nanomachines. These are synthetic DNA assemblies that switch between defined molecular shapes upon stimulation by external triggers. They can be controlled by a variety of methods that include pH changes and the addition of other molecular components, such as small molecule effectors, proteins and DNA strands. Researchers have now designed and built a simple DNA machine that is capable of continuous rotation with controlled speed and direction - a function that might be very useful for example for molecular transport. This machine is driven by an externally controlled electric field. When this field is oscillated between four directions, it continuously reorients a rotor DNA that is asymmetrically attached to a DNA axle.
 Subscribe to our Nanotechnology Spotlight feed
Subscribe to our Nanotechnology Spotlight feed





