| Posted: Oct 11, 2012 | |
Dip-pen nanolithography surface patterning with bacterial cells |
|
| (Nanowerk Spotlight) Dip-Pen Nanolithography (DPN) – developed by Chad A. Mirkin at Northwestern University in 1999 – is a scanning probe lithography technique in which the tip of an atomic force microscope (AFM) is used to 'write' molecules directly onto a substrate, allowing nanostructured surface patterning on scales of under 100 nm. DPN is the nanotechnology analog of the dip pen where the AFM tip acts as a 'pen', which is then dipped into a molecular 'ink' (i.e. coated with a chemical compound or mixture), and then used to write functional nanoscale patterns by putting it in contact with a substrate, the 'paper'. | |
| This direct-write technique offers high-resolution patterning capabilities for a number of molecular and biomolecular 'inks' on a variety of substrates, such as metals, semiconductors, and monolayer functionalized surfaces. It's becoming a work-horse tool for the scientist interested in fabricating and studying soft- and hard-matter on the sub-100nm length scale. | |
| Since the driving force of DPN for transporting materials is molecular diffusion through the sub-micrometer sized water meniscus formed between the AFM tip and the surface, large-sized ink materials are not efficiently transported through this water meniscus. For this reason, bacterial cells (1-2 micrometer or larger in length) patterning with DPN technique is often considered to be impossible. | |
| Overcoming this limitation, a Korean research team led by Jung-Hyurk Lim at Korea National University of Transportation in Chungju, has developed a 'stamp-on' DPN method that uses a previously developed hydrogel-coated tip (read more: "Dip-pen nanolithography with a porous tip generates nanopatterns with viruses") and carrier agents to generate micrometer-sized bacterial cells. | |
| In their new work, reported in the September 19, 2012 online edition of Journal of the American Chemical Society ("Direct-Write Patterning of Bacterial Cells by Dip-Pen Nanolithography"), the researchers demonstrate, for the first time, DPN patterning of large bacterial cells in a direct-write manner. | |
 |
|
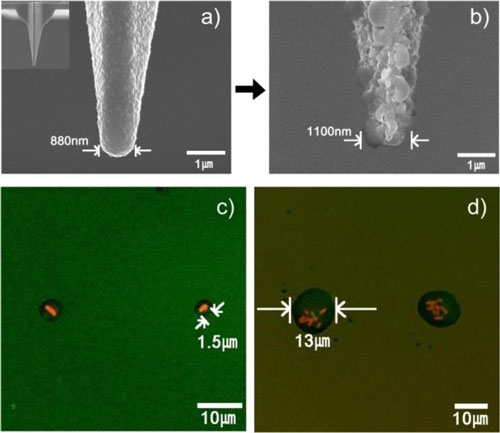
| S(a) Typical SEM image of a bare silicone oxide AFM tip. (Inset: SEM image of an entire bare tip.) (b) SEM image of a nanostructured PMeOx-coated tip (c) Fluorescence microscopy image of a pattern of individual E. coli JM 109 bacterial single cell. (c) Fluorescence microscopy image of E. coli bacterial cell (7-8 cells per one “stamp-on” feature) patterns. (Reprinted with permission from American Chemical Society) | |
| "We have shown that this approach enables the deposition of a single bacterial cell on a solid surface as well as a layering of multiple cells through a modulation of carrier solution viscosity," Lim explains to Nanowerk. "Furthermore, we also showed that a living bacterial cells array was successfully generated on an Luria-Bertani agar-coated solid surface." | |
| Lim points out that the carrier agent, such as glycerol, that was included in the ink solution could increase the mobility of the bacterial cells, and therefore it played a key role in the patterning process. "We were able to control the number of cells transported by changing the percentage of glycerol in the ink solution." | |
| As proof-of-concept, the team prepared two specialized E. coli bacteria inks to allow transport of the micrometer-sized cells from an AFM tip to a solid surface. They then successfully generated patterns of single E. coli cells on amine-functionalized silicon oxide surfaces. | |
| One ink solution contained 10% glycerol and 100 mM tricine as carrier agents, and the other included 20% glycerol with 100 mM tricine. The viscosity of the ink, which controls the number of bacterial cells transported during the patterning process, was modulated by varying the glycerol concentration. | |
| Lim says that bacterial cells could be used as power (or torque) generators for biomotors because many bacterial cells are motile. "In that kind of study, it is very important to pattern or align cells with a nanometer scale resolution. We hope that this technique could be used for those working in biological motors and machines." | |
| This novel technique will offer possibilities to biology researchers who want to generate complex arrays of bacterial cells and other microorganisms to study their interactions. | |
 By
Michael
Berger
– Michael is author of three books by the Royal Society of Chemistry:
Nano-Society: Pushing the Boundaries of Technology,
Nanotechnology: The Future is Tiny, and
Nanoengineering: The Skills and Tools Making Technology Invisible
Copyright ©
Nanowerk LLC
By
Michael
Berger
– Michael is author of three books by the Royal Society of Chemistry:
Nano-Society: Pushing the Boundaries of Technology,
Nanotechnology: The Future is Tiny, and
Nanoengineering: The Skills and Tools Making Technology Invisible
Copyright ©
Nanowerk LLC
|
|
|
Become a Spotlight guest author! Join our large and growing group of guest contributors. Have you just published a scientific paper or have other exciting developments to share with the nanotechnology community? Here is how to publish on nanowerk.com. |
|
